Characterization and Distribution of Pogonomyrmex Harvester
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Double Mound of the Harvester Ant Pogonomyrmex Occidentalis (Hymenoptera: Formicidae, Myrmicinae)
Great Basin Naturalist Volume 53 Number 4 Article 13 12-28-1993 Double mound of the harvester ant Pogonomyrmex occidentalis (Hymenoptera: Formicidae, Myrmicinae) William H. Clark Orma J. Smith Museum of Natural History, Albertson College of Idaho, Caldwell, Idaho Follow this and additional works at: https://scholarsarchive.byu.edu/gbn Recommended Citation Clark, William H. (1993) "Double mound of the harvester ant Pogonomyrmex occidentalis (Hymenoptera: Formicidae, Myrmicinae)," Great Basin Naturalist: Vol. 53 : No. 4 , Article 13. Available at: https://scholarsarchive.byu.edu/gbn/vol53/iss4/13 This Note is brought to you for free and open access by the Western North American Naturalist Publications at BYU ScholarsArchive. It has been accepted for inclusion in Great Basin Naturalist by an authorized editor of BYU ScholarsArchive. For more information, please contact [email protected], [email protected]. Creat Basin Naturalist 53(4), pp. 407-408 DOUBLE MOUND OF THE HARVESTER ANT POGONOMYRMEX OCCLDENTALIS (HYMENOPTERA: FORMICIDAE, MYRMICINAE) William H. ClarkI Key words: ho.rvester ant double mound, Pogonomyrmex occidentalis, ant nest, UUilt. Several species ofPogOlWmynnex, especial County, Utah, approximately 58 Ian NE Moab ly members of the occidentalis complex, have along Highway 128 and about 4 Ian from the nests surmounted by a large, conical mound Colorado River, at an elevation of 1445 m. of soil and gravel in a clearing created by the Voucher specimens from both mounds (WHC ants (Cole 1932, 1968). I report a rare double #8184 and 8185) are deposited in the Orma J. mound of the western halvester ant, Pogono Smith Museum of Natural History, Albertson mymwx oecidentalis (Cresson), having two College ofIdaho, Caldwell (ClDA). -

Sunshine Is Delicious, Rain Is Refreshing, Wind Braces Us Up, Snow Is Exhilarating; There Is Really No Such Thing As Bad Weather
3/14/11 The Midden Photo by Nathan Veatch Galveston Bay Area Chapter - Texas Master Naturalists April 2011 Table of Contents AT/Stewardship/Ed 2 Spring is Here by Diane Humes, President 2011 Outreach Opportunities Prairie Ponderings 3 Sunshine is delicious, rain is refreshing, wind braces us up, snow is exhilarating; there Wetland Wanderings 4 is really no such thing as bad weather, only different kinds of good weather. - John Ruskin Spring 2011 Class 6 Fishy Facts 7 We have had all kinds of good weather this winter, threat of ice, snow, and sleet unfortunately causing our first-ever chapter meeting cancellation in February. “Heartbreak” Turtles 7 Fortunately, we all stayed warm, dry, and safe. Our speaker, Chris LaChance, has re- Hook ‘em Horns 9 scheduled her presentation for the June meeting, so all is now well. We look forward Horny Toads 10 to all different kinds of good weather and good times for the coming year! FoGISP Training 11 The new chapter training class began February 17 and Raptor Workshop 12 continues most Thursdays into May. Check the schedule and please drop by to meet the 22 future Guppies from Julie 13 Galveston Bay Area Master Naturalists and welcome Red Harvester Ants 14 them to our mission of preservation, restoration, and education about our natural environment. The new class members bring a wealth of experience and love of nature, most citing experiences camping and exploring the outdoors since childhood. They are already into the food, fun, and friendship! The year 2011 marks our chapter’s 10th anniversary and plans are being formulated for a year of celebration. -

Texas Horned Lizard Watch Monitoring Packet
Management and Monitoring Packet What’s happened to contents all the horny toads? About Horned Lizards......................2 Everyone loves horny toads, but for many Texans, the fierce-looking, yet Management of amiable, reptiles are only a fond childhood memory. Once common through- out most of the state, horned lizards have disappeared from many parts of Horned Lizards .................................4 their former range. How to Monitor A statewide survey conducted by the Horned Lizard Conservation Society in Horned Lizards .................................7 1992 confirmed many Texans’ personal experiences—in the latter part of the Landowner Access 20th century the Texas Horned Lizard nearly disappeared from the eastern Request Form ...................................8 third of Texas and many respondents reported that horned lizards were in- creasingly rare in Central and North Texas. Only in West and South Texas do Site Survey Data Form .....................9 populations seem somewhat stable. Transect Data Form ...................... 10 Many factors have been proposed as culprits in the disappearance of horned lizards, often fondly called “horny toads,” including collection for the pet Contact Information ..................... 11 trade, spread of the red imported fire ant, changes in agricultural land use, habitat loss and fragmentation as a result of urbanization, and environmental References ..................................... 11 contaminants. For the most part, however, the decline of horned lizards has remained a mystery with little understanding of management actions that could be taken to reverse it. Even less is known about the status of our other two horned lizard species—the Round-tailed Horned Lizard and the Greater Short-horned Lizard. But you can help! Through participation in Texas Horned Lizard Watch as a citizen scientist, you can collect data and observations about the presence or absence of horned lizards and habitat characteristics on your monitoring site. -

Wildland Fire in Ecosystems: Effects of Fire on Fauna
United States Department of Agriculture Wildland Fire in Forest Service Rocky Mountain Ecosystems Research Station General Technical Report RMRS-GTR-42- volume 1 Effects of Fire on Fauna January 2000 Abstract _____________________________________ Smith, Jane Kapler, ed. 2000. Wildland fire in ecosystems: effects of fire on fauna. Gen. Tech. Rep. RMRS-GTR-42-vol. 1. Ogden, UT: U.S. Department of Agriculture, Forest Service, Rocky Mountain Research Station. 83 p. Fires affect animals mainly through effects on their habitat. Fires often cause short-term increases in wildlife foods that contribute to increases in populations of some animals. These increases are moderated by the animals’ ability to thrive in the altered, often simplified, structure of the postfire environment. The extent of fire effects on animal communities generally depends on the extent of change in habitat structure and species composition caused by fire. Stand-replacement fires usually cause greater changes in the faunal communities of forests than in those of grasslands. Within forests, stand- replacement fires usually alter the animal community more dramatically than understory fires. Animal species are adapted to survive the pattern of fire frequency, season, size, severity, and uniformity that characterized their habitat in presettlement times. When fire frequency increases or decreases substantially or fire severity changes from presettlement patterns, habitat for many animal species declines. Keywords: fire effects, fire management, fire regime, habitat, succession, wildlife The volumes in “The Rainbow Series” will be published during the year 2000. To order, check the box or boxes below, fill in the address form, and send to the mailing address listed below. -

Honey Bee (Apis Mellifera) Intracolonial Genetic Diversity Influences Worker Nutritional Status Bruce J
Honey bee (Apis mellifera) intracolonial genetic diversity influences worker nutritional status Bruce J. Eckholm, Ming H. Huang, Kirk E. Anderson, Brendon M. Mott, Gloria Degrandi-Hoffman To cite this version: Bruce J. Eckholm, Ming H. Huang, Kirk E. Anderson, Brendon M. Mott, Gloria Degrandi-Hoffman. Honey bee (Apis mellifera) intracolonial genetic diversity influences worker nutritional status. Api- dologie, Springer Verlag, 2015, 46 (2), pp.150-163. 10.1007/s13592-014-0311-4. hal-01284439 HAL Id: hal-01284439 https://hal.archives-ouvertes.fr/hal-01284439 Submitted on 7 Mar 2016 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Apidologie (2015) 46:150–163 Original article * INRA, DIB and Springer-Verlag France, 2014 DOI: 10.1007/s13592-014-0311-4 Honey bee (Apis mellifera ) intracolonial genetic diversity influences worker nutritional status 1 2 3 3 Bruce J. ECKHOLM , Ming H. HUANG , Kirk E. ANDERSON , Brendon M. MOTT , 3 Gloria DEGRANDI-HOFFMAN 11025 Zylstra Road, Coupeville, WA 98239, USA 2Eurofins Agroscience Services, Inc., 15250 NC Hwy 86 South, Prospect Hill, NC 27314, USA 3Carl Hayden Bee Research Center, USDA-ARS, 2000 East Allen Road, Tucson, AZ 85719, USA Received 9 January 2014 – Revised 10 July 2014 – Accepted 25 July 2014 Abstract – Honey bee queens mate with multiple males resulting in high intracolonial genetic diversity among nestmates; a reproductive strategy known as extreme polyandry. -

Pogonomyrmex Barbatus
Anita. Behav., 1986, 34, 1402-1419 The dynamics of the daily round of the harvester ant colony (Pogonomyrmex barbatus) DEBORAH M. GORDON Museum of Comparative Zoology and Society of Fellows, Harvard University, Cambridge, Massachusetts 02138, U.S.A. Abstract. Colonies of the red harvester ant, Pogonomyrmex barbatus, do various tasks outside the nest. There is a daily temporal pattern in the numbers of ants engaged in each of five activities: foraging, nest maintenance, patrolling, midden work and convening. Perturbations were carried out in the field to investigate how the daily round changes in response to environmental events and colony needs. Interfering with nest maintenance, foraging or both caused changes in the temporal patterns in all five of the observed activities. Removing nest maintenance workers, foragers or both caused the numbers involved in all five activities to decrease, and there were temporal patterns in the effects of removals. The results of both interference and removal experiments show that the extent to which a worker group does one activity affects the behaviour of other groups. When nest maintenance or foraging is impeded experimentally, these two activities are of reciprocal priority. When both are impeded, foraging is of higher priority than nest maintenance. Harvester ants forage, patrol, maintain the nest speaking, that of an equilibrium state, but the area and foraging trails, collect and arrange dynamics of colony organization are not yet under- pebbles on the nest, and gather in small groups, stood. What events will alter the daily round, by inspecting and grooming each other. The beha- how much, and for how long? Is there more than viour of the colony outside the nest at any moment one equilibrium state? can be described by citing the number of ants This study begins an exploration of the dynamics engaged in each of the five activities. -

Evolution of Colony Characteristics in the Harvester Ant Genus
Evolution of Colony Characteristics in The Harvester Ant Genus Pogonomyrmex Dissertation zur Erlangung des naturwissenschaftlichen Doktorgrades der Bayerischen Julius-Maximilians-Universität Würzburg vorgelegt von Christoph Strehl Nürnberg Würzburg 2005 - 2 - - 3 - Eingereicht am: ......................................................................................................... Mitglieder der Prüfungskommission: Vorsitzender: ............................................................................................................. Gutachter : ................................................................................................................. Gutachter : ................................................................................................................. Tag des Promotionskolloquiums: .............................................................................. Doktorurkunde ausgehändigt am: ............................................................................. - 4 - - 5 - 1. Index 1. Index................................................................................................................. 5 2. General Introduction and Thesis Outline....................................................... 7 1.1 The characteristics of an ant colony...................................................... 8 1.2 Relatedness as a major component driving the evolution of colony characteristics.................................................................................................10 1.3 The evolution -

TAXONOMIC IDENTITY of "HALLUCINOGENIC" HARVESTER ANT Lpogonomyrmex Calijornicus) CONFIRMED
Journal of Ethnobiology 21(2): 133-144 Winter 2001 TAXONOMIC IDENTITY OF "HALLUCINOGENIC" HARVESTER ANT lPogonomyrmex calijornicus) CONFIRMED KEVIN I' GROARK Department of Anthropology, Un;ocrsity of California, Los Angeles Los Angeles, CA 90024 ABSTRACf.-The use of California harvester ants (fbgollomyrmex californiClls) for visionary and therapeutic ends was an important but poorly-documented tradi tion in native south-central California. In this brief report, a confirmation of the taxonomic identity of the red ant species used in California is presented, and the descriptive record of its use is supplemented with additional ethnographic ac counts. This taxonomic identification of this species is of particular importance, as visionary red ant ingestion provides the only well-documented case of the widespread use of an insect as an hallucinogenic agent. RESUMEN.-La utilizaci6n de hormigas granivoras rojas (HJgo/wmyrmex califor nicus) con fines alucin6genos y terapeuticos, fue una tradici6n de mucha impor tancia pero mal documentada en el sur y centro-sur de California. Este breve articulo confirma la identidad taxon6mica de dicha especie y la descripci6n de su uso se hace a !raves de datos etnogrMicos adicionales. Esta identificaci6n taxo n6mica es de especial interes, puesto que es el dnico ejemplo etnografico debi damente documentado de un agente alucin6geno derivado de un insecto. REsUME.-L'u!ilisation des fourmis moissonneuses rouges (fbgonomyrmex califor nicus) a des desseins religieux et therapeutiques etait une tradition peu docu mentee mais importante dans la vie de plusieurs groupes autochtones du centre sud de la Californie. Dans ce bref expose on trouve la confirmation de I'identification taxonomique de la fourmi et a la description de la methode de son utilisation s'ajoute des donnees ethnographiques suplementaires. -

Hybridization in Ants
Rockefeller University Digital Commons @ RU Student Theses and Dissertations 2020 Hybridization in Ants Ian Butler Follow this and additional works at: https://digitalcommons.rockefeller.edu/ student_theses_and_dissertations Part of the Life Sciences Commons HYBRIDIZATION IN ANTS A Thesis Presented to the Faculty of The Rockefeller University in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy by Ian Butler June 2020 © Copyright by Ian Butler 2020 HYBRIDIZATION IN ANTS Ian Butler, Ph.D. The Rockefeller University 2020 Interspecific hybridization is a relatively common occurrence within all animal groups. Two main factors make hybridization act differently in ants than in other species: eusociality and haplodiploidy. These factors serve to reduce the costs of interspecific hybridization in ants while simultaneously allowing them to take advantage of certain benefits. Eusociality may mitigate the effects of hybridization by allowing hybrids to be shunted into the worker caste, potentially reducing the effects of hybrid sterility. In haplodiploid species, males do not have a father. They instead develop from unfertilized eggs as haploid clones of their mother. This means that interspecifically mated queens do not completely sacrifice reproductive potential even if all hybrids are sterile because they can still produce fertile males. These factors in turn suggest that hybridization should be more common among the social Hymenoptera than other animal groups. Nevertheless, current data suggest that ants hybridize at rates similar to other animal groups, although these data are limited. Furthermore, there is a large amount of overlap between cases of interspecific hybridization and cases of genetic caste determination. A majority of the cases in ants where caste is determined primarily by genotype are associated with hybridization. -

Red Harvester Ant (Big Red Ants; Harvester Ants, Red Ants; Barbatus Harvester Ant)
Pest Profile Photo credit: April Nobile, California Academy of Sciences (Specimen CASENT0006306; from https://www.antweb.org) Common Name: Red harvester ant (big red ants; harvester ants, red ants; Barbatus harvester ant) Scientific Name: Pogonomyrmex barbatus Order and Family: Order Hymenoptera; Family Formicidae Size and Appearance: Length (mm) Appearance Egg Larva/Nymph Adult Workers Pogonomyrmex harvester ants are characterized by broad, range from 5 boxy heads, 12-segmented antennae, a two-part waist, a pair mm – 8 mm of dorsal spines and a stinger. Many species have a set of long hairs (called a psammophore) on the posterior lower portion of the head, behind the mouthparts. Workers of the red harvester ant are polymorphic without distinct majors and minors. They vary in color from a light to very deep red and they have texture in the form of lines on the head (rugae). Colonies have one queen (monogynous). Queens are similar to workers but larger with a larger thorax. Female and male reproductive ants have wings. Pupa (if applicable) Type of feeder (Chewing, sucking, etc.): Chewing Host(s): Harvester ants in the genus Pogonomyrmex specialize in seeds but may forage on other foods (generalist and opportunistic). Description of Damage (larvae and adults): Because theses ants primarily eat seeds (up to 90% of the diet), they are pests of agricultural systems, damaging crops but especially grasslands (pasture). They harvest seeds, defoliate plants, and remove young and old plants. Agricultural important plants they affect include corn, oats, alfalfa, cotton, guayule, grapes, date, citrus trees, apple, and pear, as well as more generally pasture (grasses) and shrubs. -
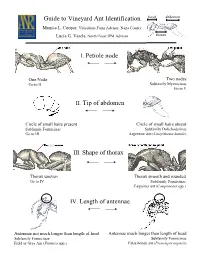
I. Petiole Node II. Tip of Abdomen IV. Length of Antennae Guide to Vineyard Ant Identification III. Shape of Thorax
Guide to Vineyard Ant Identification head abdomen Monica L. Cooper, Viticulture Farm Advisor, Napa County Lucia G. Varela, North Coast IPM Advisor thorax I. Petiole node One Node Two nodes Go to II Subfamily Myrmicinae Go to V II. Tip of abdomen Circle of small hairs present Circle of small hairs absent Subfamily Formicinae Subfamily Dolichoderinae Go to III Argentine Ant (Linepithema humile) III. Shape of thorax Thorax uneven Thorax smooth and rounded Go to IV Subfamily Formicinae Carpenter ant (Camponotus spp.) IV. Length of antennae Antennae not much longer than length of head Antennae much longer than length of head Subfamily Formicinae Subfamily Formicinae Field or Gray Ant (Formica spp.) False honey ant (Prenolepis imparis) head abdomen Petiole with two nodes Subfamily Myrmicinae (V-VIII) thorax V. Dorsal side of Thorax & Antennae One pair of spines on thorax No spines on thorax 12 segmented antennae 10 segmented antennae Go to VI Solenopsis molesta and Solenopsis xyloni VI. Underside of head No brush of bristles Brush of long bristles Go to VII Harvester ants (Pogonomyrmex californicus and P. brevispinosis) VII. Head and Thorax With hairs Without hairs Go to VIII Cardiocondyla mauritanica VIII. Head and Thorax With many parallel furrows Without parallel furrows Profile of thorax rounded Profile of thorax not evenly rounded Pavement ant (Tetramorium “species E”) Pheidole californica Argentine Ant (Linepithema humile), subfamily Dolichoderinae Exotic species 3-4 mm in length Deep brown to light black Move rapidly in distinct trails Feed on honeydew Shallow nests (2 inches from soil surface) Alex Wild Does not bite or sting Carpenter Ant (Camponotus spp.), subfamily Formicinae Large ant: >6 mm in length Dark color with smooth, rounded thorax Workers most active at dusk and night One of most abundant and widespread genera worldwide Generalist scavengers and predators: feed on dead and living insects, nectar, fruit juices and Jack K. -

Effects of Hybridization and Life History Tradeoffs on Pathogen Resistance in the Harvester Ants (Pogonomyrmex) Yainna M
University of Vermont ScholarWorks @ UVM Graduate College Dissertations and Theses Dissertations and Theses 2015 Effects of hybridization and life history tradeoffs on pathogen resistance in the Harvester ants (Pogonomyrmex) Yainna M. Hernaiz-Hernandez University of Vermont Follow this and additional works at: https://scholarworks.uvm.edu/graddis Part of the Biology Commons Recommended Citation Hernaiz-Hernandez, Yainna M., "Effects of hybridization and life history tradeoffs on pathogen resistance in the Harvester ants (Pogonomyrmex)" (2015). Graduate College Dissertations and Theses. 518. https://scholarworks.uvm.edu/graddis/518 This Thesis is brought to you for free and open access by the Dissertations and Theses at ScholarWorks @ UVM. It has been accepted for inclusion in Graduate College Dissertations and Theses by an authorized administrator of ScholarWorks @ UVM. For more information, please contact [email protected]. EFFECTS OF HYBRIDIZATION AND LIFE HISTORY TRADEOFFS ON PATHOGEN RESISTANCE IN THE HARVESTER ANTS (POGONOMY RMEX) A Thesis Presented by Yainna Hernáiz-Hernández to The Faculty of the Graduate College of The University of Vermont In Partial Fulfillment of the Requirements for the Degree of Master of Science Specializing in Biology May, 2015 Defense Date: March 26, 2015 Thesis Examination Committee: Sara Helms Cahan, Ph.D., Advisor Douglas Johnson, Ph.D., Chairperson Joseph Schall, Ph.D., Emeriti Faculty Cynthia J. Forehand, Ph.D., Dean of the Graduate College ABSTRACT A fundamental challenge faced by all organisms is the risk of infection by pathogens that can significantly reduce their fitness. The evolutionary dynamic between hosts and pathogens is expected to be a coevolutionary cycle, as pathogens evolve by increasing their level of virulence and hosts respond by increasing their level of resistance.