THE HUMAN CELL ATLAS White Paper
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

RECENT ADVANCES in BIOLOGY, BIOPHYSICS, BIOENGINEERING and COMPUTATIONAL CHEMISTRY
RECENT ADVANCES in BIOLOGY, BIOPHYSICS, BIOENGINEERING and COMPUTATIONAL CHEMISTRY Proceedings of the 5th WSEAS International Conference on CELLULAR and MOLECULAR BIOLOGY, BIOPHYSICS and BIOENGINEERING (BIO '09) Proceedings of the 3rd WSEAS International Conference on COMPUTATIONAL CHEMISTRY (COMPUCHEM '09) Puerto De La Cruz, Tenerife, Canary Islands, Spain December 14-16, 2009 Recent Advances in Biology and Biomedicine A Series of Reference Books and Textbooks Published by WSEAS Press ISSN: 1790-5125 www.wseas.org ISBN: 978-960-474-141-0 RECENT ADVANCES in BIOLOGY, BIOPHYSICS, BIOENGINEERING and COMPUTATIONAL CHEMISTRY Proceedings of the 5th WSEAS International Conference on CELLULAR and MOLECULAR BIOLOGY, BIOPHYSICS and BIOENGINEERING (BIO '09) Proceedings of the 3rd WSEAS International Conference on COMPUTATIONAL CHEMISTRY (COMPUCHEM '09) Puerto De La Cruz, Tenerife, Canary Islands, Spain December 14-16, 2009 Recent Advances in Biology and Biomedicine A Series of Reference Books and Textbooks Published by WSEAS Press www.wseas.org Copyright © 2009, by WSEAS Press All the copyright of the present book belongs to the World Scientific and Engineering Academy and Society Press. All rights reserved. No part of this publication may be reproduced, stored in a retrieval system, or transmitted in any form or by any means, electronic, mechanical, photocopying, recording, or otherwise, without the prior written permission of the Editor of World Scientific and Engineering Academy and Society Press. All papers of the present volume were peer reviewed -

Applied Category Theory for Genomics – an Initiative
Applied Category Theory for Genomics { An Initiative Yanying Wu1,2 1Centre for Neural Circuits and Behaviour, University of Oxford, UK 2Department of Physiology, Anatomy and Genetics, University of Oxford, UK 06 Sept, 2020 Abstract The ultimate secret of all lives on earth is hidden in their genomes { a totality of DNA sequences. We currently know the whole genome sequence of many organisms, while our understanding of the genome architecture on a systematic level remains rudimentary. Applied category theory opens a promising way to integrate the humongous amount of heterogeneous informations in genomics, to advance our knowledge regarding genome organization, and to provide us with a deep and holistic view of our own genomes. In this work we explain why applied category theory carries such a hope, and we move on to show how it could actually do so, albeit in baby steps. The manuscript intends to be readable to both mathematicians and biologists, therefore no prior knowledge is required from either side. arXiv:2009.02822v1 [q-bio.GN] 6 Sep 2020 1 Introduction DNA, the genetic material of all living beings on this planet, holds the secret of life. The complete set of DNA sequences in an organism constitutes its genome { the blueprint and instruction manual of that organism, be it a human or fly [1]. Therefore, genomics, which studies the contents and meaning of genomes, has been standing in the central stage of scientific research since its birth. The twentieth century witnessed three milestones of genomics research [1]. It began with the discovery of Mendel's laws of inheritance [2], sparked a climax in the middle with the reveal of DNA double helix structure [3], and ended with the accomplishment of a first draft of complete human genome sequences [4]. -

FINALIST DIRECTORY VIRTUAL REGENERON ISEF 2020 Animal Sciences
FINALIST DIRECTORY VIRTUAL REGENERON ISEF 2020 Animal Sciences ANIM001 Dispersal and Behavior Patterns between ANIM010 The Study of Anasa tristis Elimination Using Dispersing Wolves and Pack Wolves in Northern Household Products Minnesota Carter McGaha, 15, Freshman, Vici Public Schools, Marcy Ferriere, 18, Senior, Cloquet, Senior High Vici, OK School, Cloquet, MN ANIM011 The Ketogenic Diet Ameliorates the Effects of ANIM002 Antsel and Gretal Caffeine in Seizure Susceptible Drosophila Avneesh Saravanapavan, 14, Freshman, West Port melanogaster High School, Ocala, FL Katherine St George, 17, Senior, John F. Kennedy High School, Bellmore, NY ANIM003 Year Three: Evaluating the Effects of Bifidobacterium infantis Compared with ANIM012 Development and Application of Attractants and Fumagillin on the Honeybee Gut Parasite Controlled-release Microcapsules for the Nosema ceranae and Overall Gut Microbiota Control of an Important Economic Pest: Flower # Varun Madan, 15, Sophomore, Lake Highland Thrips, Frankliniella intonsa Preparatory School, Orlando, FL Chunyi Wei, 16, Sophomore, The Affiliated High School of Fujian Normal University, Fuzhou, ANIM004T Using Protease-activated Receptors (PARs) in Fujian, China Caenorhabditis elegans as a Potential Therapeutic Agent for Inflammatory Diseases ANIM013 The Impacts of Brandt's Voles (Lasiopodomys Swetha Velayutham, 15, Sophomore, brandtii) on the Growth of Plantations Vyshnavi Poruri, 15, Sophomore, Surrounding their Patched Burrow Units Plano East, Senior High School, Plano, TX Meiqi Sun, 18, Senior, -

Abstracts Genome 10K & Genome Science 29 Aug - 1 Sept 2017 Norwich Research Park, Norwich, Uk
Genome 10K c ABSTRACTS GENOME 10K & GENOME SCIENCE 29 AUG - 1 SEPT 2017 NORWICH RESEARCH PARK, NORWICH, UK Genome 10K c 48 KEYNOTE SPEAKERS ............................................................................................................................... 1 Dr Adam Phillippy: Towards the gapless assembly of complete vertebrate genomes .................... 1 Prof Kathy Belov: Saving the Tasmanian devil from extinction ......................................................... 1 Prof Peter Holland: Homeobox genes and animal evolution: from duplication to divergence ........ 2 Dr Hilary Burton: Genomics in healthcare: the challenges of complexity .......................................... 2 INVITED SPEAKERS ................................................................................................................................. 3 Vertebrate Genomics ........................................................................................................................... 3 Alex Cagan: Comparative genomics of animal domestication .......................................................... 3 Plant Genomics .................................................................................................................................... 4 Ksenia Krasileva: Evolution of plant Immune receptors ..................................................................... 4 Andrea Harper: Using Associative Transcriptomics to predict tolerance to ash dieback disease in European ash trees ............................................................................................................ -

Single-Cell Transcriptome Analysis As a Promising Tool to Study Pluripotent Stem Cell Reprogramming
International Journal of Molecular Sciences Review Single-Cell Transcriptome Analysis as a Promising Tool to Study Pluripotent Stem Cell Reprogramming Hyun Kyu Kim †, Tae Won Ha † and Man Ryul Lee * Soonchunhyang Institute of Medi-Bio Science (SIMS), Soon Chun Hyang University, Cheonan 31151, Korea; [email protected] (H.K.K.); [email protected] (T.W.H.) * Correspondence: [email protected]; Tel.: +81-41-413-5013 † These authors equally contributed to this work. Abstract: Cells are the basic units of all organisms and are involved in all vital activities, such as proliferation, differentiation, senescence, and apoptosis. A human body consists of more than 30 trillion cells generated through repeated division and differentiation from a single-cell fertilized egg in a highly organized programmatic fashion. Since the recent formation of the Human Cell Atlas consortium, establishing the Human Cell Atlas at the single-cell level has been an ongoing activity with the goal of understanding the mechanisms underlying diseases and vital cellular activities at the level of the single cell. In particular, transcriptome analysis of embryonic stem cells at the single-cell level is of great importance, as these cells are responsible for determining cell fate. Here, we review single-cell analysis techniques that have been actively used in recent years, introduce the single-cell analysis studies currently in progress in pluripotent stem cells and reprogramming, and forecast future studies. Keywords: single-cell mRNA sequencing; pluripotent stem cell; somatic cell reprogramming; in- duced pluripotent stem cell; heterogeneity Citation: Kim, H.K.; Ha, T.W.; Lee, M.R. Single-Cell Transcriptome Analysis as a Promising Tool to Study Pluripotent Stem Cell Reprogramming. -

Introduction to the Cell Cell History Cell Structures and Functions
Introduction to the cell cell history cell structures and functions CK-12 Foundation December 16, 2009 CK-12 Foundation is a non-profit organization with a mission to reduce the cost of textbook materials for the K-12 market both in the U.S. and worldwide. Using an open-content, web-based collaborative model termed the “FlexBook,” CK-12 intends to pioneer the generation and distribution of high quality educational content that will serve both as core text as well as provide an adaptive environment for learning. Copyright ©2009 CK-12 Foundation This work is licensed under the Creative Commons Attribution-Share Alike 3.0 United States License. To view a copy of this license, visit http://creativecommons.org/licenses/by-sa/3.0/us/ or send a letter to Creative Commons, 171 Second Street, Suite 300, San Francisco, California, 94105, USA. Contents 1 Cell structure and function dec 16 5 1.1 Lesson 3.1: Introduction to Cells .................................. 5 3 www.ck12.org www.ck12.org 4 Chapter 1 Cell structure and function dec 16 1.1 Lesson 3.1: Introduction to Cells Lesson Objectives • Identify the scientists that first observed cells. • Outline the importance of microscopes in the discovery of cells. • Summarize what the cell theory proposes. • Identify the limitations on cell size. • Identify the four parts common to all cells. • Compare prokaryotic and eukaryotic cells. Introduction Knowing the make up of cells and how cells work is necessary to all of the biological sciences. Learning about the similarities and differences between cell types is particularly important to the fields of cell biology and molecular biology. -

PBMC Fixation and Processing for Chromium Single-Cell RNA Sequencing
bioRxiv preprint doi: https://doi.org/10.1101/315267; this version posted May 5, 2018. The copyright holder has placed this preprint (which was not certified by peer review) in the Public Domain. It is no longer restricted by copyright. Anyone can legally share, reuse, remix, or adapt this material for any purpose without crediting the original authors. PBMC Fixation and Processing for Chromium Single-Cell RNA Sequencing Jinguo Chen1, Foo Cheung1, Rongye Shi1,2, Huizhi Zhou1,2, Wenrui Lu1,3, CHI Consortium1 1Center for Human Immunology, Autoimmunity and Inflammation (CHI), National Institutes of Health 2Department of Transfusion Medicine, Clinical Center, National Institutes of Health 3Fujian Provincial Hospital, Fuzhou, China CHI Consortium (Julián Candia1, Yuri Kotliarov1, Katie R. Stagliano1 and John S. Tsang1,2) 1 CHI 2 Systems Genomics and Bioinformatics Unit, NIAID Abstract Background: Interest in single-cell transcriptomic analysis is growing rapidly, especially for profiling rare or heterogeneous populations of cells. In almost all reported works, investigators have used live cells which introduces cell stress during preparation and hinders complex study designs. Recent studies have indicated that cells fixed by denaturing fixative can be used in single-cell sequencing. But they did not work with most primary cells including immune cells. Methods: The methanol-fixation and new processing method was introduced to preserve PBMCs for single-cell RNA sequencing (scRNA-Seq) analysis on 10X Chromium platform. Results: When methanol fixation protocol was broken up into three steps: fixation, storage and rehydration, we found that PBMC RNA was degraded during rehydration with PBS, not at cell fixation and up to three-month storage steps. -
Creating a Census of Human Cells
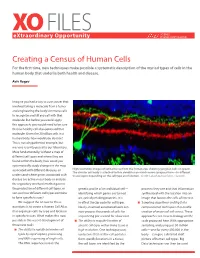
XO FILES SCIENCE eXtraordinary Opportunity PHILANTHROPY ALLIANCE Creating a Census of Human Cells For the first time, new techniques make possible a systematic description of the myriad types of cells in the human body that underlie both health and disease. Aviv Regev Imagine you had a way to cure cancer that involved taking a molecule from a tumor and engineering the body’s immune cells to recognize and kill any cell with that molecule. But before you could apply this approach, you would need to be sure that no healthy cell also expressed that molecule. Given the 20 trillion cells in a human body, how would you do that? This is not a hypothetical example, but was one recently posed to our laboratory. More fundamentally, without a map of different cell types and where they are found within the body, how could you systematically study changes in the map associated with different diseases, or High resolution images of retinal tissue from the human eye, showing ganglion cells (in green). The circular cell body is attached to thin dendrites on which nerve synapses form—in different understand where genes associated with tissue layers depending on the cell type and function. Credit: Lab of Joshua Sanes, Harvard. disease are active in our body or analyze the regulatory mechanisms that govern the production of different cell types, or genetic profile of an individual cell— proteins they use and that information sort out how different cell types combine identifying which genes are turned synthesized with the location into an to form specific tissues? on, actively making proteins. -

Microblogging the ISMB: a New Approach to Conference Reporting
Message from ISCB Microblogging the ISMB: A New Approach to Conference Reporting Neil Saunders1*, Pedro Beltra˜o2, Lars Jensen3, Daniel Jurczak4, Roland Krause5, Michael Kuhn6, Shirley Wu7 1 School of Molecular and Microbial Sciences, University of Queensland, St. Lucia, Brisbane, Queensland, Australia, 2 Department of Cellular and Molecular Pharmacology, University of California San Francisco, San Francisco, California, United States of America, 3 Novo Nordisk Foundation Center for Protein Research, Panum Institute, Copenhagen, Denmark, 4 Department of Bioinformatics, University of Applied Sciences, Hagenberg, Freistadt, Austria, 5 Max-Planck-Institute for Molecular Genetics, Berlin, Germany, 6 European Molecular Biology Laboratory, Heidelberg, Germany, 7 Stanford Medical Informatics, Stanford University, Stanford, California, United States of America Cameron Neylon entitled FriendFeed for Claire Fraser-Liggett opened the meeting Scientists: What, Why, and How? (http:// with a review of metagenomics and an blog.openwetware.org/scienceintheopen/ introduction to the human microbiome 2008/06/12/friendfeed-for-scientists-what- project (http://friendfeed.com/search?q = why-and-how/) for an introduction. room%3Aismb-2008+microbiome+OR+ We—a group of science bloggers, most fraser). The subsequent Q&A session of whom met in person for the first time at covered many of the exciting challenges The International Conference on Intel- ISMB 2008—found FriendFeed a remark- for those working in this field. Clearly, ligent Systems for Molecular Biology -

Xiaowei Zhuang Harvard University, Howard Hughes Medical Institute Xiaowei Zhuang Is the David B
Xiaowei Zhuang Harvard University, Howard Hughes Medical Institute Xiaowei Zhuang is the David B. Arnold Professor of Science at Harvard University and an investigator of Howard Hughes Medical Institute. Her laboratory has developed single-molecule, super-resolution and genomic- scale imaging methods, including STORM and MERFISH, and has used these methods to discover novel molecular structures in cells and cell organizations in tissues. Zhuang received her BS in physics from the University of Science and Technology of China, her PhD in physics in the lab of Prof. Y. R. Shen at University of California, Berkeley, and her postdoctoral training in biophysics in the lab of Prof. Steven Chu at Stanford University. She joined the faculty of Harvard University in 2001 and became a Howard Hughes Medical Institute investigator in 2005. Zhuang is a member of the US National Academy of Sciences and the American Academy of Arts and Sciences, a foreign member of the Chinese Academy of Sciences and the European Molecular Biology Organization, a fellow of the American Association of the Advancement of Science and the American Physical Society. She received honorary doctorate degrees from the Stockholm University in Sweden and the Delft University of Technology in the Netherlands. She has received a number of awards, including the Breakthrough Prize in Life Sciences, National Academy of Sciences Award in Scientific Discovery, Dr. H.P. Heineken Prize for Biochemistry and Biophysics, National Academy of Sciences Award in Molecular Biology, Raymond and Beverly Sackler International Prize in Biophysics, Max Delbruck Prize in Biological Physics, American Chemical Society Pure Chemistry Award, MacArthur Fellowship, etc. -

Robustness and Timing of Cellular Differentiation Through Population-Based Symmetry Breaking
AR TI CLE Robustness AND TIMING OF CELLULAR DIFFERENTIATION THROUGH population-based SYMMETRY BREAKING Angel StanoeV1 | Christian Schröter1 | Aneta KOSESKA1∗ 1Department OF Systemic Cell Biology, Max Planck Institute OF Molecular PhYSIOLOGY, Although COORDINATED BEHAVIOR OF MULTICELLULAR ORGANISMS Dortmund, GermanY RELIES ON INTERCELLULAR communication, THE DYNAMICAL mech- Correspondence ANISM THAT UNDERLIES SYMMETRY BREAKING IN HOMOGENEOUS Email: populations, THEREBY GIVING RISE TO HETEROGENEOUS CELL TYPES ∗[email protected] REMAINS UNCLEAR. In CONTRAST TO THE PREVALENT cell-intrinsic VIEW OF DIFFERENTIATION WHERE MULTISTABILITY ON THE SINGLE CELL LEVEL IS A NECESSARY pre-requisite, WE PROPOSE THAT het- EROGENEOUS CELLULAR IDENTITIES EMERGE AND ARE MAINTAINED COOPERATIVELY ON A POPULATION LEVel. Identifying A GENERIC SYMMETRY BREAKING mechanism, WE DEMONSTRATE THAT ROBUST PROPORTIONS BETWEEN DIFFERENTIATED CELL TYPES ARE INTRINSIC FEATURES OF THIS INHOMOGENEOUS STEADY STATE solution. Crit- ICAL ORGANIZATION IN THE VICINITY OF THE SYMMETRY BREAKING BIFURCATION ON THE OTHER hand, SUGGESTS THAT TIMING OF cel- LULAR DIFFERENTIATION EMERGES FOR GROWING POPULATIONS IN A self-organized MANNER. Setting A CLEAR DISTINCTION BETWEEN pre-eXISTING ASYMMETRIES AND SYMMETRY BREAKING EVents, WE THUS DEMONSTRATE THAT EMERGENT SYMMETRY breaking, IN CONJUNCTION WITH ORGANIZATION NEAR CRITICALITY NECESSARILY LEADS TO ROBUSTNESS AND ACCURACY DURING DEVelopment. KEYWORDS SYMMETRY breaking; differentiation; INHOMOGENEOUS STEADY STATE Abbreviations: -

Research Focuses on the Gen- Maureen R
This Month Surveying resident physician exposure to COVID-19 2 Alternative mechanisms underlying acute graft- versus-host disease 4 Targeting melanocortin signaling alleviates cachexia 5 Intrathecal gene therapy improves neuromuscular deterioration in canine model 6 September 2020 JCI This Month is a summary of the most recent articles in The Journal of Clinical Investigation and JCI Insight jci.org/this-month Placental infection by SARS–CoV-2 p. 2 Scan for the digital version of JCI This Month. Journal of Clinical Investigation Consulting Editors Soman N. Abraham Richard T. D'Aquila Katherine A. High Terri Laufer David J. Pinsky M. Celeste Simon John S. Adams Alan Daugherty Helen H. Hobbs Mitchell A. Lazar Edward Plow Mihaela Skobe Qais Al-Awqati Sudhansu Dey Ronald Hoffman Brendan Lee Catherine Postic Donald Small Kari Alitalo Anna Mae Diehl V. Michael Holers William M.F. Lee Alice S. Prince Lois Smith Dario C. Altieri Harry C. Dietz III Steven Holland Rudolph L. Leibel Louis J. Ptáček Akrit Sodhi Masayuki Amagai Gianpietro Dotti David Holtzman Wayne I. Lencer Luigi Puglielli Weihong Song Brian H. Annex Michael Dustin Michael J. Holtzman Jon D. Levine Pere Puigserver Ashley L. St. John M. Amin Arnaout Connie J. Eaves Lawrence B. Holzman Ross L. Levine Bali Pulendran Jonathan Stamler Alan Attie Dominique Eladari Tamas L. Horvath Klaus Ley Ellen Puré Colin L. Stewart Jane E. Aubin Joel K. Elmquist Gokhan S. Hotamisligil Rodger A. Liddle Susan E. Quaggin Doris Stoffers Michael F. Beers Stephen G. Emerson Steven R. Houser Richard Locksley Marlene Rabinovitch Warren Strober Vann Bennett Jonathan A.