A Bibliometric Insight of Genetic Factors in ASD: Emerging Trends and New Developments
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Do Neuroscience Journals Accept Replications? a Survey of Literature
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by HKU Scholars Hub Do Neuroscience Journals Accept Replications? A Survey of Title Literature Author(s) Yeung, WKA Citation Frontiers in Human Neuroscience, 2017, v. 11, p. 468 Issued Date 2017 URL http://hdl.handle.net/10722/247231 This Document is Protected by copyright and was first published by Frontiers. All rights reserved. It is reproduced with Rights permission.; This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License. ORIGINAL RESEARCH published: 20 September 2017 doi: 10.3389/fnhum.2017.00468 Do Neuroscience Journals Accept Replications? A Survey of Literature Andy W. K. Yeung* Oral and Maxillofacial Radiology, Applied Oral Sciences, Faculty of Dentistry, The University of Hong Kong, Hong Kong, Hong Kong Background: Recent reports in neuroscience, especially those concerning brain-injury and neuroimaging, have revealed low reproducibility of results within the field and urged for more replication studies. However, it is unclear if the neuroscience journals welcome or discourage the submission of reports on replication studies. Therefore, the current study assessed the explicit position of neuroscience journals on replications. Methods: A list of active neuroscience journals publishing in English was compiled from Scopus database. These journal websites were accessed to read their aims and scope and instructions to authors, and to assess if they: (1) explicitly stated that they accept replications; (2) did not state their position on replications; (3) implicitly discouraged replications by emphasizing on the novelty of the manuscripts; or (4) explicitly stated that they reject replications. -

Central Nervous System Manifestations in COVID‐19 Patients
Received: 30 July 2020 | Revised: 23 November 2020 | Accepted: 20 December 2020 DOI: 10.1002/brb3.2025 REVIEW Central nervous system manifestations in COVID-19 patients: A systematic review and meta-analysis Shahrzad Nazari1 | Amirhossein Azari Jafari2 | Seyyedmohammadsadeq Mirmoeeni2 | Saeid Sadeghian3 | Mohammad Eghbal Heidari4 | Siavash Sadeghian5 | Farhad Assarzadegan6 | Seyed Mahmoud Puormand7 | Hamid Ebadi8 | Davood Fathi9,10 | Sahar Dalvand11 1Department of Neuroscience and Addiction Studies, School of Advanced Technologies in Medicine, Tehran University of Medical Sciences, Tehran, Iran 2Student Research Committee, School of Medicine, Shahroud University of Medical Sciences, Shahroud, Iran 3Department of Paediatric Neurology, Golestan Medical, Educational, and Research Centre, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran 4Students Scientific Research Center, Tehran University Medical Science, Tehran, Iran 5Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran 6Department of Neurology, Imam Hossein Hospital, Shahid Beheshti University of Medical Sciences, Tehran, Iran 7Imam Hossein Hospital, Shahid Beheshti University of Medical Sciences, Tehran, Iran 8Department of Clinical Neurosciences, University of Calgary, Calgary, AB, Canada 9Brain and Spinal Cord Injury Research Center, Neuroscience Institute, Tehran University of Medical Sciences, Tehran, Iran 10Department of Neurology, Shariati Hospital, Tehran University of Medical Sciences, Tehran, Iran 11Functional Neurosurgery Research Center, Shohada Tajrish Comprehensive Neurosurgical Center of Excellence, Shahid Beheshti University of Medical Sciences, Tehran, Iran Correspondence Sahar Dalvand, Functional Neurosurgery Abstract Research Center, Shohada Tajrish Background: At the end of December 2019, a novel respiratory infection, initially Comprehensive Neurosurgical Center of Excellence, Shahid Beheshti University of reported in China, known as COVID-19 initially reported in China, and later known Medical Sciences, Tehran, Iran. as COVID-19, led to a global pandemic. -

Print Special Issue Flyer
IMPACT FACTOR 3.394 an Open Access Journal by MDPI Longitudinal Assessment of Alcohol Exposure on Brain and Behavior Guest Editors: Message from the Guest Editors Prof. Dr. Cheryl L Kirstein Alcoholism is a complex disease that typically begins with Department of Psychology, alcohol experimentation and use early in development. University of South Florida, Tampa, FL 33620, USA Early childhood and early to late adolescence have been shown to be critical periods during which the developing [email protected] brain is particularly susceptible to alcohol’s effects. Alcohol Prof. Dr. Antoinette administration during development has been shown to Maldonado-Devincci facilitate adulthood dependence. Importantly, Department of Psychology, North behavioral/personality traits have been utilized to Carolina Agricultural and Technical State University, determine predictability of escalation to increased alcohol Greensboro, NC 27411, USA use and abuse. Recent studies have begun to examine this [email protected] important issue from a developmental perspective in both males and females. Alcohol addiction is a progressive disease that does not typically begin in adults, rather during a time of rapid brain development. Investigating Deadline for manuscript and addressing the impact of alcohol and the submissions: closed (25 February 2021) developmental progression of addiction, is paramount to understanding how to mitigate alcohol’s impact on the brain (molecular to behavioral mechanisms) and, ultimately, society. mdpi.com/si/34304 SpeciaIslsue IMPACT FACTOR 3.394 an Open Access Journal by MDPI Editor-in-Chief Message from the Editor-in-Chief Prof. Dr. Stephen D. Meriney You are invited to contribute a research article or a Department of Neuroscience, comprehensive review for consideration and publication in University of Pittsburgh, Brain Sciences (ISSN 2076-3425). -

Brain and Language
BRAIN AND LANGUAGE AUTHOR INFORMATION PACK TABLE OF CONTENTS XXX . • Description p.1 • Impact Factor p.1 • Abstracting and Indexing p.1 • Editorial Board p.2 • Guide for Authors p.3 ISSN: 0093-934X DESCRIPTION . Aims and Scope An interdisciplinary journal, Brain and Language publishes articles that elucidate the complex relationships among language, brain, and behavior. The journal covers the large variety of modern techniques in cognitive neuroscience, including functional and structural brain imaging, electrophysiology, cellular and molecular neurobiology, genetics, lesion-based approaches, and computational modeling. All articles must relate to human language and be relevant to the understanding of its neurobiological and neurocognitive bases. Published articles in the journal are expected to have significant theoretical novelty and/or practical implications, and use perspectives and methods from psychology, linguistics, and neuroscience along with brain data and brain measures. Benefits to authors We also provide many author benefits, such as free PDFs, a liberal copyright policy, special discounts on Elsevier publications and much more. Please click here for more information on our author services. Please see our Guide for Authors for information on article submission. If you require any further information or help, please visit our Support Center IMPACT FACTOR . 2020: 2.381 © Clarivate Analytics Journal Citation Reports 2021 ABSTRACTING AND INDEXING . Scopus PubMed/Medline Science Citation Index Embase CSA Neurosciences Abstracts -

Global Research Trends in Microbiome-Gut-Brain Axis During 2009–2018
Zyoud et al. BMC Gastroenterology (2019) 19:158 https://doi.org/10.1186/s12876-019-1076-z RESEARCHARTICLE Open Access Global research trends in microbiome-gut- brain axis during 2009–2018: a bibliometric and visualized study Sa’ed H. Zyoud1,2,3* , Simon Smale4, W. Stephen Waring5, Waleed M. Sweileh6 and Samah W. Al-Jabi2 Abstract Background: The pathways and mechanism by which associations between the gut microbiome and the brain, termed the microbiome-gut-brain axis (MGBA), are manifest but remain to be fully elucidated. This study aims to use bibliometric analysis to estimate the global activity within this rapidly developing field and to identify particular areas of focus that are of current relevance to the MGBA during the last decade (2009–2018). Methods: The current study uses the Scopus for data collection. We used the key terms “microbiome-gut-brain axis” and its synonyms because we are concerned with MGBA per se as a new concept in research rather than related topics. A VOSviewer version 1.6.11 was used to visualize collaboration pattern between countries and authors, and evolving research topics by analysis of the term co-occurrence in the title and abstract of publications. Results: Between 2009 and 2018, there were 51,504 published documents related to the microbiome, including 1713 articles related to the MGBA: 829 (48.4%) original articles, 658(38.4%) reviews, and 226 (13.2%) other articles such as notes, editorials or letters. The USA took the first place with 385 appearances, followed by Ireland (n =161),China(n = 155), and Canada (n = 144).The overall citation h-index was 106, and the countries with the highest h-index values were the USA (69), Ireland (58), and Canada (43). -
HORMONES and BEHAVIOR Official Journal of the Society for Behavioral Neuroendocrinology
HORMONES AND BEHAVIOR Official Journal of the Society for Behavioral Neuroendocrinology AUTHOR INFORMATION PACK TABLE OF CONTENTS XXX . • Description p.1 • Impact Factor p.1 • Abstracting and Indexing p.1 • Editorial Board p.2 • Guide for Authors p.5 ISSN: 0018-506X DESCRIPTION . Hormones and Behavior publishes original research articles, reviews, commentaries, perspectives and special issues concerning hormone-brain-behavior relationships, broadly defined. The journal welcomes studies conducted on species ranging from invertebrates to mammals, including studies of humans. Also included is work addressing neuroendocrine-behavior relationships, neuroendocrine and endocrine mechanisms controlling the development or adult expression of behavior as well as studies of the environmental control and evolutionary significance of hormone-brain-behavior relationships. Studies with an emphasis on molecular, neuroanatomical, or non-neural systems are also welcome when relevant to endocrine signaling and behavior. Benefits to authors We also provide many author benefits, such as free PDFs, a liberal copyright policy, special discounts on Elsevier publications and much more. Please click here for more information on our author services. Please see our Guide for Authors for information on article submission. If you require any further information or help, please visit our Support Center IMPACT FACTOR . 2020: 3.587 © Clarivate Analytics Journal Citation Reports 2021 ABSTRACTING AND INDEXING . Scopus CrossRef EBSCOhost Embase ProQuest PubMed/Medline PsycINFO Web of Science Science Citation Index Expanded Social Sciences Citation Index AUTHOR INFORMATION PACK 23 Sep 2021 www.elsevier.com/locate/yhbeh 1 EDITORIAL BOARD . Editor C.M. McCormick, Brock University, St Catharines, L2S 3A1, Ontario, Canada Associate Editors J. Balthazart, University of Liege, Liege, Belgium J.M. -

Print Special Issue Flyer
IMPACT FACTOR 3.394 an Open Access Journal by MDPI Cannabis: Neuropsychiatry and Its Effects on Brain and Behavior Guest Editors: Message from the Guest Editors Dr. Marco Colizzi Over the years, there has been increasing interest into the public Section of Psychiatry, health impact of cannabis use. Recreational cannabis use, Department of Neurosciences, Biomedicine and Movement especially a frequent use of products with high levels of its main Sciences, University of Verona, psychoactive ingredient delta-9-tetrahydrocannabinol (Δ 9-THC), Verona, Italy; can cause dependence and have transient and long-lasting Department of Psychosis Studies, Institute of Psychiatry, detrimental mental health effects, also negatively impacting Psychology & Neuroscience, cognitive processing and brain function and metabolism. In King's College London, London, regular users, the development of tolerance to some of the effects UK of cannabis, especially the pleasurable ones, may lead to [email protected] progressively heavier use in order to obtain the same effects in terms of their intensity, with higher health risks. Dr. Sagnik Bhattacharyya Department of Psychosis Studies, Different cannabinoids may modulate human brain function and Institute of Psychiatry, Psychology & Neuroscience, behavior in different ways, with different risk–benefit profiles. King's College London, London, Original research articles advancing our understanding of the United Kingdom “Neuropsychiatry and Brain effects of Cannabis” are solicited for sagnik.2.bhattacharyya@ this Special Issue. Reviews providing an analytical perspective on kcl.ac.uk the existing literature are also welcome. Deadline for manuscript submissions: closed (15 May 2020) mdpi.com/si/25478 SpeciaIslsue IMPACT FACTOR 3.394 an Open Access Journal by MDPI Editor-in-Chief Message from the Editor-in-Chief Prof. -

Neuroepigenetics
NEUROEPIGENETICS AUTHOR INFORMATION PACK TABLE OF CONTENTS XXX . • Description p.1 • Abstracting and Indexing p.1 • Editorial Board p.1 • Guide for Authors p.3 ISSN: 2214-7845 DESCRIPTION . Neuroepigenetics is a brand new, open access journal from Elsevier which aims to provide a forum for cross-disciplinary research exploring how epigenetic mechanisms contribute to brain health and disease across the lifespan, transgenerationally and on an evolutionary scale. The journal will publish original research articles, review articles, position papers, methods articles and commentaries on technological issues related to research tools and data analysis, articles highlighting meeting proceedings. The journal welcomes research investigating epigenetic mechanisms underlying brain structure and function, cognition and behavior at all levels of biological complexity, studies of epigenetic mechanisms mediating environmental influences on brain and behavior as well as research exploring the therapeutic and diagnostic potential of epigenetic mechanisms. Also of interest is research integrating epigenomic with genomic, gene expression and other “omic” data to gain mechanistic insight about brain function inhealth and disease. This latest addition to Elseviers Neurology portfolio will be an online, open access journal. There will be no need to subscribe or to register to read published articles. Authors will retain copyright, and reuse will be permitted by means of creative commons licensing. As such an article processing fee will apply; however, in the journals initial stages significant discounts will be offered. Find out more and submit your paper at: http://ees.elsevier.com/NEPIG. Elsevier's high standards of peer review and editing will apply. ABSTRACTING AND INDEXING . Directory of Open Access Journals (DOAJ) EDITORIAL BOARD . -

Author Guidelines
--------------------------------- Version 16-March-2012 Author Guidelines Did you know... Journal of Neurochemistry has no page charges? Instructions to Authors During submission, authors will be asked to indicate: Full affiliations (salutation, first and last (family) name, institution, department, city, email address) of the corresponding author Salutation, first and last name, institution, department, city of all co-authors Title, abstract, up to 6 keywords for the manuscript An area the manuscript fits in (Bioenergetics & Metabolism, Brain Development & Cell Differentiation, Genetics & Gene Regulation, Molecular Basis of Disease, Neuroinflammation & Neuroimmunology, Neuronal Plasticity & Behavior, Signal Transduction & Synaptic Transmission) Names of preferred referees (full name and email address) and handling editors; while this option is not mandatory, suggestions are welcomed Full name and email address of non-preferred referees, if needed Cover letter to the Editor Details of whether the manuscript is a resubmission (if yes, you will need to provide the previous manuscript ID) If the manuscript has colour figures If the first or last author is younger than age 35 (for the manuscript to be eligible for the Mark A. Smith Award) If you are an ISN member All manuscript files: o Main text file including figure and table legends as .doc or .rtf o Tables (may also be included at the end of the main text file) o Figures as .tiff, .eps or .pdf (one file per figure) with at least 300 dpi resolution Alternatively, a single .ps -
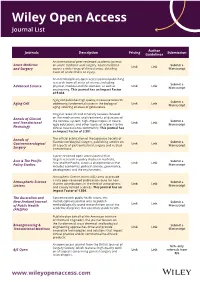
Wiley Open Access Journal List
Wiley Open Access Journal List Author Journals Description Pricing Submission Guidelines An international peer-reviewed academic journal Acute Medicine on acute medicine and surgery. Acute medicine Submit a Link Link and Surgery covers a wide range of clinical areas, detailing Manuscript cases of acute illness or injury. An interdisciplinary open access journal publishing research from all areas of science, including Submit a physical, medical and life sciences, as well as Link Link Advanced Science Manuscript engineering. This journal has an Impact Factor of 9.034. Aging Cell publishes high quality, innovative research Submit a addressing fundamental issues in the biology of Link Link Aging Cell Manuscript aging, covering all areas of geroscience. Original research and scholarly reviews focused Annals of Clinical on the mechanisms and treatments of diseases of the nervous system; high-impact topics in neuro- Submit a Link Link and Translational logic education; and other topics of interest to the Manuscript Neurology clinical neuroscience community. This journal has an Impact Factor of 3.901. Annals of The official publication of the Japanese Society of Gastroenterological Surgery, publishing articles on Submit a Link Link Gastroenterological all aspects of gastrointestinal surgery and related Manuscript Surgery interventions. A peer-reviewed open access journal that targets research in policy studies in Australia, Asia & The Pacific Submit a Asia and the Pacific, across a discipline focus that Link Link Manuscript Policy Studies includes economics, political science, governance, development and the environment. Atmospheric Science Letters (ASL) aims to provide a fully peer-reviewed publication route for new Atmospheric Science Submit a shorter contributions in the field of atmospheric Link Link Manuscript Letters and closely related sciences. -

Download the Author Guidelines As A
As of October 18, 2019, all manuscripts are now submitted through the Research Exchange (REX) Submission Portal. For submissions started prior to October 18, 2019, including revised submissions or invited submissions, please visit ScholarOne to manage your submission. If you have any questions about the submission portals, please email [email protected] or [email protected]. Editorial Process Overview Journal Aim and Scope The Journal of Neuroscience Research (JNR) publishes novel research results that will advance our understanding of the development, function and pathophysiology of the nervous system, using molecular, cellular, systems, and translational approaches. The journal focuses on uncovering the intricacies of brain structure and function. Research published in JNR covers all species from invertebrates to humans, and the reports inform the readers about the function and organization of the nervous system, with emphasis on how disease modifies the function and organization. The acceptance criteria for all papers are the quality and originality of the research, and its significance to the journal’s readership. Except where otherwise stated, manuscripts are single-blind peer reviewed. Papers will only be sent to review if the Editor-in-Chief determines that the paper meets the appropriate quality and relevance requirements. The Journal does not consider Case Reports or studies using herbal remedies/natural compounds derived from plants as these are difficult to reproduce. In addition, studies involving immortalized cell lines that are not accompanied by primary cultures, slice or in vivo work will not be considered as it is difficult or impossible to extrapolate results to normal neuronal populations. Peer Review Peer review by two to three external reviewers will be made after editorial prescreening.