(12) Patent Application Publication (10) Pub. No.: US 2005/0221316A1 Pedersen Et Al
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

9Th & 10Th Grade
1/27/2019 SpeechWire Tournament Services Ridge Invitational Declamation (9th & 10th Grade) Elimination round results Final round All Judges Code Name Team LK1 WK1 AA1 All ranks Place recips pref. AH101 Niamh Campbell Phoenix Country Day School 1 1 6 14 6.67 1 LP103 Matthew Floyd Catholic Memorial 2 2 1 15 5.25 2 BD109 Ashin Varghese Randolph High School 3 3 2 16 5.42 3 AO121 Juliette Gringeri Summit High School 5 4 4 23 3.95 4 UT102 Farrah Culmer Democracy Prep Harlem Prep 7 6 3 24 4.48 5 AH108 Enzo Acharya Phoenix Country Day School 6 5 7 26 4.01 6 AG101 Brynn Nelson Fontbonne Hall Academy 4 7 5 26 3.84 7 Semifinal round All Judges Code Name Team AP2 BD1 All ranks Place recips pref. AH101 Niamh Campbell Phoenix Country Day School 1 1 6 4.50 1 BD109 Ashin Varghese Randolph High School 8 4.25 2 UT102 Farrah Culmer Democracy Prep Harlem Prep 8 3.83 3 AH108 Enzo Acharya Phoenix Country Day School 8 3.50 4 AG101 Brynn Nelson Fontbonne Hall Academy 10 3.25 5 AO121 Juliette Gringeri Summit High School 10 3.25 5 LP103 Matthew Floyd Catholic Memorial 2 2 10 3.25 5 AG102 Juliann Bianco Fontbonne Hall Academy 11 3.53 8 AL101 Adrian Tejada Democracy Prep Endurance High 11 3.00 9 AH112 Milan Sewell Phoenix Country Day School 12 2.58 10 BC115 Maria Chabanov Xaverian HS 13 2.08 11 AH109 Devan Amin Phoenix Country Day School 14 2.37 12 GG105 Uswad Qureshi Phillipsburg HS 14 2.33 13 AC107 Nina Li Strath Haven High School 3 4 14 1.92 14 BC116 Amy Gruber Xaverian HS 15 2.70 15 AH133 Ella Brenes Phoenix Country Day School 4 5 15 2.28 16 AM101 Jaina Jallow Livingston High School 5 3 15 1.87 17 AK108 Fatimata Ly Achievement First 17 2.60 18 CZ112 Rajit Sharma Union Catholic 17 2.03 19 Semifinal round All Judges Code Name Team AA5 RY3 All ranks Place recips pref. -

List 237 FINAL JOE.Vp
STEPHEN ALBUM World Coins Specializing in Oriental Numismatics Post Office Box 7386, Santa Rosa, Calif. 95407, U.S.A. 237 Telephone 707-539-2120 — Fax 707-539-3348 [email protected] Catalog price $6.00 www.stevealbum.com September 2008 Ancient Gold Coins 66505. ABBASID: al-Mansur, 754-775, AV dinar (4.24g), NM, AH140, A-212, bold strike, lovely ef $385 67809. BOSPOROS: Eupator, 155-171, AV stater (7.72g), Anokhin-542a, with Roman emperors Marcus Aurelius & Lucius Verus, dated BE461 (=164/5 AD), diademed & draped bust of Eupator right / laureate & draped bust of 66507. ABBASID: al-Mahdi, 775-785, AV dinar (4.24g), NM, Marcus Aurelius vis-a-vis bare head of Lucius Verus; spear AH158, A-214, nice strike, choice ef $350 between, date below, vf-ef, R $800 67184. ACHAIMENIDIAN EMPIRE: Uncertain King, ca. 485-420 BC, 68241. ABBASID: al-Muttaqi, 940-944, AV dinar (4.39g), Madinat AV daric (8.36g), Sardes, Carradice Type IIib A/B, al-Salam, AH331, A-260, citing the Hamdanid brothers, Nasir Persian king or hero running right, holding transverse al-Dawla & Sayf al-Dawla, inscribed ibriz (“pure gold”), vf, S $360 spear and bow / irregular rectangular incuse, lustrous ef $2,100 66503. ABBASID: al-Rashid, 786-809, AV dinar (4.25g), NM (Egypt), AH184, A-218.11, citing Ja’far, lovely strike, choice au $400 56313. HEPHTHALITE: Khingila, ca. 430-490, AV scyphate stater (7.38g), G-84, king standing, facing right, holding trident with curved handle / very crude remnants of Siva & bull, as is typical of this type, ef, R $685 23827. -
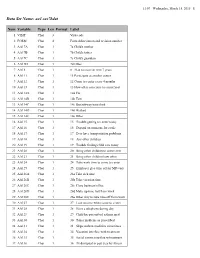
Childhood Asthma Management Program Data Dictionary
11:07 Wednesday, March 18, 2015 1 Data Set Name: aa1.sas7bdat Num Variable Type Len Format Label 1 VISIT Char 3 Visit code 2 FORM Char 4 Form abbreviation and revision number 3 AA17A Char 1 7a Child's mother 4 AA17B Char 1 7b Child's father 5 AA17C Char 1 7c Child's guardian 6 AA17D Char 1 7d Other 7 AA18 Char 1 8 Plan to move in next 7 years 8 AA111 Char 1 11 Participate at another center 9 AA112 Char 1 12 Come to center every 4 months 10 AA113 Char 1 13 How often can come to center/year 11 AA114A Char 1 14a Car 12 AA114B Char 1 14b Taxi 13 AA114C Char 1 14c Bus/subway/metrolink 14 AA114D Char 1 14d Walked 15 AA114E Char 1 14e Other 16 AA115 Char 1 15 Trouble getting to center today 17 AA116 Char 1 16 Depend on someone for a ride 18 AA117 Char 1 17 Ever have transportation problems 19 AA118 Char 1 18 Any other children 20 AA119 Char 1 19 Trouble finding child care today 21 AA120 Char 1 20 Bring other children to center ever 22 AA121 Char 1 21 Bring other children how often 23 AA124 Char 1 24 Take work time to come to center 24 AA125 Char 1 25 Employer give time off for MD visit 25 AA126A Char 1 26a Take sick time 26 AA126B Char 1 26b Take vacation time 27 AA126C Char 1 26c Close business/office 28 AA126D Char 1 26d Make up time lost from work 29 AA126E Char 1 26e Other way to take time off from work 30 AA127 Char 1 27 Lose income when come to center 31 AA128 Char 1 28 Have a telephone during day 32 AA129 Char 1 29 Child has prescribed asthma med 33 AA130 Char 1 30 Takes medicine as prescribed 34 AA131 Char 1 31 Skips asthma medicine -

Before the Public Service Commission of the State of Missouri
BEFORE THE PUBLIC SERVICE COMMISSION OF THE STATE OF MISSOURI In the Matter of Kansas City Power & Light ) Company’s Notice of Intent to File an ) Application for Authority to Establish a Demand- ) File No. EO-2015-0240 Side Programs Investment Mechanism ) In the Matter of KCP&L Greater Missouri Operations ) Company’s Notice of Intent to File an ) Application for Authority to Establish a Demand- ) File No. EO-2015-0241 Side Programs Investment Mechanism ) RESPONSE TO ORDER APPROVING APPLICATION TO MODIFY TECHNICAL RESOURCE MANUAL AND PROGRAM DESIGN INCENTIVE RANGES COMES NOW Kansas City Power & Light Company and KCP&L Greater Missouri Operations Company (the “Company”) and, in response Ordered Paragraph 2 of the Missouri Public Service Commission’s (“Commission”) March 21, 2018 Order Approving Application to Modify Technical Resource Manual and Program Design Incentive Ranges (“Order”), files the attached amended Appendix 1 and Appendix 2 (collectively referred to as “Amended Appendices”) to the Non-Unanimous Stipulation and Agreement (originally approved by the Commission on April 6, 2016). The Amended Appendices incorporate all modifications approved by the Commission in its Order. WHEREFORE, the Company respectfully requests the Commission consider this response to the Commission’s Order. Respectfully submitted, /s/ Roger W. Steiner Robert J. Hack MBN 36496 Roger W. Steiner MBN 39586 Kansas City Power & Light Company 1200 Main Street, 16th Floor Kansas City, MO 64105 (816) 556-2785 (Phone) (816) 556-2787 (Fax) [email protected] [email protected] James M. Fischer MBN 27543 Fischer & Dority, P.C. 101 Madison Street, Suite 400 Jefferson City, MO 65101 (573) 636-6758 (Phone) (573) 636-0383 (Fax) [email protected] Attorneys for Kansas City Power & Light Company and KCP&L Greater Missouri Operations Company CERTIFICATE OF SERVICE I do hereby certify that a true and correct copy of the foregoing document has been hand delivered, emailed or mailed, postage prepaid, this 26th day of March, 2018, to all counsel of record. -

Technical Assistance Layout with Instructions
Resettlement Plan August 2017 INO: Flood Management in Selected River Basins Sector Project Prepared by the Ministry of Public Works and Housing through the Directorate General of Water Resources for the Asian Development Bank. This is an updated and revised version of the draft originally posted in May 2015 available on http://www.adb.org/projects/35182-043/main#project- documents. CURRENCY EQUIVALENTS (as of 11 August 2016) Rp1.00 = $0.000076 $1.00 = Rp13,129 ABBREVIATIONS ADB – Asian Development Bank AH – affected household AP – affected person BAPPEDA – Badan Perencanaan Pembangunan Daerah (Provincial/District Development Planning Agency) BPN – Badan Pertanahan Nasional (National Land Agency) BWS – Balai Wilayah Sungai (Center for River Basin) CBFRM – community-based flood risk management COI – corridor of impact CWZ – construction works zone DED – detailed engineering design DGWR – Directorate General of Water Resources DMS – detailed measurement survey EA – executing agency EIA – environmental impact assessment EMA – External Monitoring Agency FMSRBSP – Flood Management in Selected River Basins Sector Project GOI – Government of Indonesia HH – household IA – Implementing agency IOL – inventory of losses IR – involuntary resettlement Km – kilometer LA – Land acquisition LAIT – land acquisition implementation team LRP – livelihood restoration program MAPPI – Masyarakat Profesi Penilai Indonesia (Indonesian Professional Appraiser Association) MOHA – Ministry of Home Affairs NJOP – Nilai Jual Object Pajal (tax object selling price) PIB -
Thailand Mega Project for GMS Connectivity the 9Th GMSARN International Conference 2014
Thailand Mega Project for GMS connectivity The 9th GMSARN International Conference 2014 Athibhu Chitranukroh Office of Transport and Traffic Policy and Planning Ministry of Transport, Thailand November 12th , 2014 Palace Hotel Saigon, Ho Chi Minh City, Vietnam Agenda : Thailand Mega Project for GMS connectivity Part 1: Trend of the future - Globalization & Global value chain - Urbanization - Critical Factor Part 2: Basic Fact of the GMS Part 3: Thailand - Logistics challenges - Infrastructure development program - Key projects Part 4 : Impact to the regional economy Trend : Globalization & Global Value Chain Effects of McDonalds and Starbuck’s franchises on global trade Source : The McGraw Center for teaching and learning Trend : Globalization & Global Value Chain : iPhone Trend : Globalization & Global Value Chain : Boeing 787 Trend : urbanization • 50% global GDP generated by 600 cities • Yr. 2025 : 40% global GDP will be generated by emerging markets Trend : urbanization Urban population • Yr. 1900 : 2 of 10 people live in urban • Yr. 2010 : 5 of 10 people live in urban • Yr. 2030 : 6 of 10 people live in urban • Yr. 2050 : 7 of 10 people live in urban Social : • Lack of jobs -> crime • Pollution -> disease • Traffic -> quality of life Environment : cities consume • 2/3 global energy • 60% water • CO2 70% Trend : urbanization Better Urbanization leads to higher-quality growth for all people - Urban & Transport infrastructure Trend : urbanization Trend : critical factor - Globalization & Global Value Chain - urbanization “Connectivity” -

Standard Recode Manual for DHS 6
STANDARD RECODE MANUAL FOR DHS 6 Demographic and Health Surveys Methodology This document is part of the Demographic and Health Survey’s DHS Toolkit of methodology for the MEASURE DHS Phase III project, implemented from 2008-2013. This publication was produced for review by the United States Agency for International Development (USAID). It was prepared by MEASURE DHS/ICF International. [THIS PAGE IS INTENTIONALLY BLANK] Description of the Demographic and Health Surveys Individual Recode Data File DHS VI Version 1.0 (With differences from DHS V) March 22, 2013 Foreword DHS surveys collect primary data using several types of questionnaires. A household questionnaire is used to collect information on characteristics of the household's dwelling unit, and data related to the height and weight for women and children in the household. It is also used to identify members of the household who are eligible for an individual interview. Eligible respondents are then interviewed using an individual questionnaire. In a majority of DHS surveys eligible individuals include women of reproductive age (15-49) and men age 15-59, or in some cases 15-54. In some countries only women are interviewed. Individual questionnaires include information on fertility, family planning and maternal and child health. Data are available from DHS for each of these surveys by request through the mail or from our web site at www.measuredhs.com. Data from DHS surveys are produced in both raw and recode formats. A raw data file includes the data as they were collected, without any structural changes. These files are generally not distributed, but they are also available on request. -

HQ Ful-Grip Sheaves-Single Groove for Both Belt Sections "A" (4L) and "O" (3L) D.D
HQ Ful-Grip Sheaves-Single Groove For Both Belt Sections "A" (4L) and "O" (3L) D.D. D.D. Original Dimensions - Inches O.D. A Belts O Belts Part Maurey Style Weight Inches Inches Inches Number Number (Lbs) F L P C X G 3.05 2.8 2.5 AK30H --- 1 1.1 3/4 1 1/4 7/8 --- 1-3/16 7/16 3.25 3.0 2.7 AK32H --- 1 1.2 3/4 1 1/4 7/8 --- 1-3/16 7/16 3.45 3.2 2.9 AK34H --- 2 1.0 3/4 1 1/4 9/16 --- 7/8 7/16 3.75 3.5 3.2 AK39H --- 2 1.4 3/4 1 1/4 9/16 --- 7/8 7/16 3.95 3.7 3.4 AK41H AH40 2 1.6 3/4 1 1/4 9/16 --- 7/8 7/16 4.25 4.0 3.7 AK44H AH43 2 1.9 3/4 1 1/4 9/16 --- 7/8 7/16 4.45 4.2 3.9 AK46H AH45 2 1.9 3/4 1 1/4 9/16 --- 7/8 7/16 4.75 4.5 4.2 AK49H AH48 2 2.1 3/4 1 1/4 9/16 --- 7/8 7/16 4.95 4.7 4.4 AK51H AH50 2 2.3 3/4 1 1/4 9/16 --- 7/8 7/16 5.25 5.0 4.7 AK54H AH53 2 2.0 3/4 1 1/4 9/16 --- 7/8 7/16 5.45 5.2 4.9 AK56H AH55 2 2.3 3/4 1 1/4 9/16 --- 7/8 7/16 5.75 5.5 5.2 AK59H --- 2 2.4 3/4 1 1/4 9/16 --- 7/8 7/16 5.95 5.7 5.4 AK61H AH60 4 2.5 3/4 1 1/4 9/16 0 7/8 7/16 6.25 6.0 5.7 AK64H AH63 4 2.7 3/4 1 1/4 9/16 0 7/8 7/16 6.45 6.2 5.9 AK66H AH65 4 2.8 3/4 1 1/4 9/16 0 7/8 7/16 6.75 6.5 6.2 AK69H AH68 4 2.9 3/4 1 1/4 9/16 0 7/8 7/16 6.95 6.7 6.6 AK71H AH70 4 3.1 3/4 1 1/4 9/16 0 7/8 7/16 7.25 7.0 6.7 AK74H AH73 4 3.3 3/4 1 1/4 9/16 0 7/8 7/16 7.75 7.5 7.2 AK79H AH78 4 3.5 3/4 1 1/4 9/16 0 7/8 7/16 7.95 7.7 7.4 AH80 --- 4 2.9 3/4 1 1/4 9/16 0 7/8 7/16 8.25 8.0 7.7 AK84H AH83 4 3.6 3/4 1 1/4 9/16 0 7/8 7/16 8.75 8.5 8.2 AK89H AH88 4 4.0 3/4 1 1/4 9/16 0 7/8 7/16 8.95 8.7 8.4 AH90 --- 4 2.8 3/4 1 1/4 9/16 0 7/8 7/16 9.25 9.0 8.7 AK94H AH93 4 4.4 3/4 1 -

The Maximum Diversity Assortment Selection Problem
Takustr. 7 Zuse Institute Berlin 14195 Berlin Germany FELIX PRAUSE1,KAI HOPPMANN-BAUM2,BORIS DEFOURNY3,THORSTEN KOCH4 The Maximum Diversity Assortment Selection Problem 1 0000-0001-9401-3707 2 0000-0001-9184-8215 3 0000-0003-0405-5538 4 0000-0002-1967-0077 ZIB Report 20-34 (Dezember 2020) Zuse Institute Berlin Takustr. 7 14195 Berlin Germany Telephone: +49 30-84185-0 Telefax: +49 30-84185-125 E-mail: [email protected] URL: http://www.zib.de ZIB-Report (Print) ISSN 1438-0064 ZIB-Report (Internet) ISSN 2192-7782 The Maximum Diversity Assortment Selection Problem Felix Prause1[0000−0001−9401−3707], Kai Hoppmann-Baum1;2[0000−0001−9184−8215], Boris Defourny3[0000−0003−0405−5538], and Thorsten Koch1;2[0000−0002−1967−0077] 1 Zuse Institute Berlin, Takustr. 7, 14195 Berlin, Germany {prause,hoppmann-baum,koch}@zib.de 2 TU Berlin, Chair of Software and Algorithms for Discrete Optimization, Str. des 17. Juni 135, 10623 Berlin, Germany 3 Lehigh University, Department of Industrial and Systems Engineering, 200 W Packer Ave, Bethlehem, PA, 18015, USA [email protected] Abstract. In this paper, we introduce the Maximum Diversity Assort- ment Selection Problem (MADASS), which is a generalization of the 2-dimensional Cutting Stock Problem (2CSP). Given a set of rectan- gles and a rectangular container, the goal of 2CSP is to determine a subset of rectangles that can be placed in the container without overlap- ping, i.e., a feasible assortment, such that a maximum area is covered. In MADASS, we need to determine a set of feasible assortments, each of them covering a certain minimum threshold of the container, such that the diversity among them is maximized. -

CHIS 2020 Adult CATI Questionnaire (Interviewer- Administered) Version 1.16 April 19, 2021 Adult Respondents Age 18 and Older
CHIS 2020 Adult CATI Questionnaire (Interviewer- administered) Version 1.16 April 19, 2021 Adult Respondents Age 18 and Older Collaborating Agencies: • UCLA Center for Health Policy Research • California Department of Health Care Services • California Department of Public Health Contact: California Health Interview Survey UCLA Center for Health Policy Research 10960 Wilshire Blvd, Suite 1550 Los Angeles, CA 90024 Telephone: (866) 275-2447 Fax: (310) 794-2686 Web: www.chis.ucla.edu Copyright © 2020 by the Regents of the University of California CHIS 2020 Adult Questionnaire Version 1.16 April 19, 2021 Table of Contents Section A: Demographic Information, Part I ......................................................................................... 6 Age ......................................................................................................................................................... 6 Gender Identity ...................................................................................................................................... 8 Ethnicity ................................................................................................................................................. 9 Race ..................................................................................................................................................... 10 Language Spoken at Home ................................................................................................................. 15 Additional Language Use .................................................................................................................... -

CHIS 2021 Adult CAWI Questionnaire (Self- Administered) Version 1.28 May 17, 2021 Adult Respondents Age 18 and Older
CHIS 2021 Adult CAWI Questionnaire (Self- administered) Version 1.28 May 17, 2021 Adult Respondents Age 18 and Older Collaborating Agencies: • UCLA Center for Health Policy Research • California Department of Health Care Services • California Department of Public Health Contact: California Health Interview Survey UCLA Center for Health Policy Research 10960 Wilshire Blvd, Suite 1550 Los Angeles, CA 90024 Telephone: (866) 275-2447 Fax: (310) 794-2686 Web: www.chis.ucla.edu Copyright © 2021 by the Regents of the University of California CHIS 2021 Adult Questionnaire Version 1.28 May 17, 2021 Table of Contents Section A: Demographic Information, Part I ......................................................................................... 7 Age ......................................................................................................................................................... 7 Gender Identity ...................................................................................................................................... 8 Ethnicity ................................................................................................................................................. 9 Race ..................................................................................................................................................... 10 Language Spoken at Home ................................................................................................................. 15 Additional Language Use ....................................................................................................................