H+-Proton-Pumping Inorganic Pyrophosphatase
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Identification and Partial Characterization of a Family of Putative Palmitoyltransferases in Dictyostelium Discoideum Brent Elliot Wells
The University of Maine DigitalCommons@UMaine Electronic Theses and Dissertations Fogler Library 2003 Identification and Partial Characterization of a Family of Putative Palmitoyltransferases in Dictyostelium Discoideum Brent Elliot Wells Follow this and additional works at: http://digitalcommons.library.umaine.edu/etd Part of the Biochemistry Commons Recommended Citation Wells, Brent Elliot, "Identification and Partial Characterization of a Family of Putative Palmitoyltransferases in Dictyostelium Discoideum" (2003). Electronic Theses and Dissertations. 301. http://digitalcommons.library.umaine.edu/etd/301 This Open-Access Thesis is brought to you for free and open access by DigitalCommons@UMaine. It has been accepted for inclusion in Electronic Theses and Dissertations by an authorized administrator of DigitalCommons@UMaine. IDENTIFICATION AND PARTIAL CHARACTERIZATION OF A FAMILY OF PUTATIVE PALMITOYLTRANSFERASES IN DICTYOSTELIUM DISCOIDEUM BY Brent Elliott Wells B.S. University of Utah, 1994 A THESIS Submitted in Partial Fulfillment of the Requirements for the Degree of Master of Science (in Biochemistry) The Graduate School The University of Maine December, 2003 Advisory Committee: Robert E. Gundersen, Associate Professor of Biochemistry, Advisor Keith W. Hutchison, Professor of Biochemistry Mary Rumpho-Kennedy, Professor of Biochemistry LIBRARY RIGHTS STATEMENT In presenting this thesis in partial fulfillment of the requirements for an advanced degree at The University of Maine, I agree that the Library shall make it freely available for inspection. I further agree that permission for "fair use" copying of this thesis for scholarly purposes may be granted by the Librarian. It is understood that any copying or publication of this thesis for financial gain shall not be allowed without my written permission. -

Structural Basis for the Sheddase Function of Human Meprin Β Metalloproteinase at the Plasma Membrane
Structural basis for the sheddase function of human meprin β metalloproteinase at the plasma membrane Joan L. Arolasa, Claudia Broderb, Tamara Jeffersonb, Tibisay Guevaraa, Erwin E. Sterchic, Wolfram Boded, Walter Stöckere, Christoph Becker-Paulyb, and F. Xavier Gomis-Rütha,1 aProteolysis Laboratory, Department of Structural Biology, Molecular Biology Institute of Barcelona, Consejo Superior de Investigaciones Cientificas, Barcelona Science Park, E-08028 Barcelona, Spain; bInstitute of Biochemistry, Unit for Degradomics of the Protease Web, University of Kiel, D-24118 Kiel, Germany; cInstitute of Biochemistry and Molecular Medicine, University of Berne, CH-3012 Berne, Switzerland; dArbeitsgruppe Proteinaseforschung, Max-Planck-Institute für Biochemie, D-82152 Planegg-Martinsried, Germany; and eInstitute of Zoology, Cell and Matrix Biology, Johannes Gutenberg-University, D-55128 Mainz, Germany Edited by Brian W. Matthews, University of Oregon, Eugene, OR, and approved August 22, 2012 (received for review June 29, 2012) Ectodomain shedding at the cell surface is a major mechanism to proteolysis” step within the membrane (1). This is the case for the regulate the extracellular and circulatory concentration or the processing of Notch ligand Delta1 and of APP, both carried out by activities of signaling proteins at the plasma membrane. Human γ-secretase after action of an α/β-secretase (11), and for signal- meprin β is a 145-kDa disulfide-linked homodimeric multidomain peptide peptidase, which removes remnants of the secretory pro- type-I membrane metallopeptidase that sheds membrane-bound tein translocation from the endoplasmic membrane (13). cytokines and growth factors, thereby contributing to inflammatory Recently, human meprin β (Mβ) was found to specifically pro- diseases, angiogenesis, and tumor progression. -

Inorganic Pyrophosphatase Is a Component of the Drosophila Nucleosome Remodeling Factor Complex
Downloaded from genesdev.cshlp.org on October 3, 2021 - Published by Cold Spring Harbor Laboratory Press Inorganic pyrophosphatase is a component of the Drosophila nucleosome remodeling factor complex David A. Gdula, Raphael Sandaltzopoulos, Toshio Tsukiyama,1 Vincent Ossipow, and Carl Wu2 Laboratory of Molecular Cell Biology, National Cancer Institute, National Institutes of Health, Bethesda, Maryland 20892-4255 USA The Drosophila nucleosome remodeling factor (NURF) is a protein complex consisting of four polypeptides that facilitates the perturbation of chromatin structure in vitro in an ATP-dependent manner. The 140-kD NURF subunit, imitation switch (ISWI), is related to the SWI2/SNF2 ATPase. Another subunit, NURF-55, is a 55-kD WD repeat protein homologous to the human retinoblastoma-associated protein RbAp48. Here, we report the cloning and characterization of the smallest (38 kD) component of NURF. NURF-38 is strikingly homologous to known inorganic pyrophosphatases. Both recombinant NURF-38 alone and the purified NURF complex are shown to have inorganic pyrophosphatase activity. Inhibition of the pyrophosphatase activity of NURF with sodium fluoride has no significant effect on chromatin remodeling, indicating that these two activities may be biochemically uncoupled. Our results suggest that NURF-38 may serve a structural or regulatory role in the complex. Alternatively, because accumulation of unhydrolyzed pyrophosphate during nucleotide incorporation inhibits polymerization, NURF may also have been adapted to deliver pyrophosphatase -

Protein Family Classification with Neural Networks
Protein Family Classification with Neural Networks Timothy K. Lee Tuan Nguyen Program in Biomedical Informatics Department of Statistics Stanford University Stanford University [email protected] [email protected] Abstract Understanding protein function from amino acid sequence is a fundamental prob- lem in biology. In this project, we explore how well we can represent biological function through examination of raw sequence alone. Using a large corpus of protein sequences and their annotated protein families, we learn dense vector rep- resentations for amino acid sequences using the co-occurrence statistics of short fragments. Then, using this representation, we experiment with several neural net- work architectures to train classifiers for protein family identification. We show good performance for a multi-class prediction problem with 589 protein family classes. 1 Introduction Next-generation sequencing technologies generate large amounts of biological sequence informa- tion in the form of DNA/RNA sequences. From DNA sequences we also know the amino acid sequences of proteins, which are the fundamental molecules that perform most biological functions. The functionality of a protein is thus encoded in the amino acid sequence and understanding the sequence-function relationship is a major challenge in bioinformatics. Investigating protein func- tional often involves structural studies (crystallography) or biochemical studies, which require time consuming efforts. Protein families are defined to group together proteins that share similar function, and the aim of our project is to predict protein family from raw sequence. We focus on training infor- mative vector representations for protein sequences and investigate various neural network models for the task of predicting a protein’s family. -

Pyruvate-Phosphate Dikinase of Oxymonads and Parabasalia and the Evolution of Pyrophosphate-Dependent Glycolysis in Anaerobic Eukaryotes† Claudio H
EUKARYOTIC CELL, Jan. 2006, p. 148–154 Vol. 5, No. 1 1535-9778/06/$08.00ϩ0 doi:10.1128/EC.5.1.148–154.2006 Copyright © 2006, American Society for Microbiology. All Rights Reserved. Pyruvate-Phosphate Dikinase of Oxymonads and Parabasalia and the Evolution of Pyrophosphate-Dependent Glycolysis in Anaerobic Eukaryotes† Claudio H. Slamovits and Patrick J. Keeling* Canadian Institute for Advanced Research, Botany Department, University of British Columbia, 3529-6270 University Boulevard, Vancouver, British Columbia V6T 1Z4, Canada Received 29 September 2005/Accepted 8 November 2005 In pyrophosphate-dependent glycolysis, the ATP/ADP-dependent enzymes phosphofructokinase (PFK) and pyruvate kinase are replaced by the pyrophosphate-dependent PFK and pyruvate phosphate dikinase (PPDK), respectively. This variant of glycolysis is widespread among bacteria, but it also occurs in a few parasitic anaerobic eukaryotes such as Giardia and Entamoeba spp. We sequenced two genes for PPDK from the amitochondriate oxymonad Streblomastix strix and found evidence for PPDK in Trichomonas vaginalis and other parabasalia, where this enzyme was thought to be absent. The Streblomastix and Giardia genes may be related to one another, but those of Entamoeba and perhaps Trichomonas are distinct and more closely related to bacterial homologues. These findings suggest that pyrophosphate-dependent glycolysis is more widespread in eukaryotes than previously thought, enzymes from the pathway coexists with ATP-dependent more often than previously thought and may be spread by lateral transfer of genes for pyrophosphate-dependent enzymes from bacteria. Adaptation to anaerobic metabolism is a complex process (PPDK), respectively (for a comparison of these reactions, see involving changes to many proteins and pathways of critical reference 21). -

Gent Forms of Metalloproteinases in Hydra
Cell Research (2002); 12(3-4):163-176 http://www.cell-research.com REVIEW Structure, expression, and developmental function of early diver- gent forms of metalloproteinases in Hydra 1 2 3 4 MICHAEL P SARRAS JR , LI YAN , ALEXEY LEONTOVICH , JIN SONG ZHANG 1 Department of Anatomy and Cell Biology University of Kansas Medical Center Kansas City, Kansas 66160- 7400, USA 2 Centocor, Malvern, PA 19355, USA 3 Department of Experimental Pathology, Mayo Clinic, Rochester, MN 55904, USA 4 Pharmaceutical Chemistry, University of Kansas, Lawrence, KS 66047, USA ABSTRACT Metalloproteinases have a critical role in a broad spectrum of cellular processes ranging from the breakdown of extracellular matrix to the processing of signal transduction-related proteins. These hydro- lytic functions underlie a variety of mechanisms related to developmental processes as well as disease states. Structural analysis of metalloproteinases from both invertebrate and vertebrate species indicates that these enzymes are highly conserved and arose early during metazoan evolution. In this regard, studies from various laboratories have reported that a number of classes of metalloproteinases are found in hydra, a member of Cnidaria, the second oldest of existing animal phyla. These studies demonstrate that the hydra genome contains at least three classes of metalloproteinases to include members of the 1) astacin class, 2) matrix metalloproteinase class, and 3) neprilysin class. Functional studies indicate that these metalloproteinases play diverse and important roles in hydra morphogenesis and cell differentiation as well as specialized functions in adult polyps. This article will review the structure, expression, and function of these metalloproteinases in hydra. Key words: Hydra, metalloproteinases, development, astacin, matrix metalloproteinases, endothelin. -

Annotation of Glycolysis and Gluconeogenesis Pathways
A metabolic insight into the Asian citrus psyllid: Annotation of glycolysis and gluconeogenesis pathways Blessy Tamayo, Kyle Kercher, Tom D’Elia, Helen Wiersma-Koch, Surya Saha, Teresa Shippy, Susan J. Brown, and Prashant Hosmani Introduction Glycolysis, as in other animals, is the major metabolic pathway that insects use to extract energy from carbohydrates [1]. This process consists of ten reactions that convert one molecule of glucose into two molecules of pyruvate within the cytosol, generating a net gain of two molecules of ATP. Simple carbohydrates are acquired through the insect diet and processed through glycolysis and the central carbohydrate metabolic steps. Comparative genomic analysis of Apis mellifera, Drosophila melanogaster, and Anopheles gambiae has revealed the presence of genes for metabolic pathways that support dietary habits dependent on sugar-rich substrates ([2]; [3]; [4]; [5]). In addition to processing sugars for energy, glycolysis also serves as a central pathway that serves as both a source of important precursors and destination for key intermediates from many metabolic pathways. Gluconeogenesis is the process through which glucose is synthesized from non- carbohydrate substrates, and is closely associated with glycolysis. Eleven enzymatic reactions occur during gluconeogenesis. Eight of the enzymes involved in the steps also catalyze the reverse reactions in glycolysis and the three remaining enzymes are specific to gluconeogenesis. 1 Gluconeogenesis generated carbohydrates are required as substrate for anaerobic glycolysis, synthesis of chitin, glycoproteins, polyols and glycoside detoxication products [1]. Gluconeogenesis is essential in insects to maintain sugar homeostasis and serves as the initial process towards the generation of glucose disaccharide trehalose, which is the main circulating sugar in the insect hemolymph ([6]; [7]). -
Type of the Paper (Article
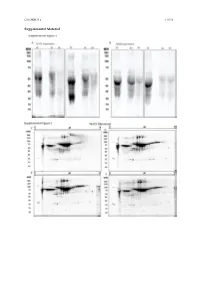
Cells 2020, 9, x 1 of 19 Supplemental Material Cells 2020, 9, x 2 of 19 Figure S1. Secretome enrichment: protocol optimization. 1D SDS-PAGE documentation of washing steps: Culture medium was substituted with FCS-free medium, which was changed every 2 h. The supernatants were then collected, and the proteins isolated and separated in 1D SDS-PAGE ((A) TK173 and (B) TK188). Proteins were stained with Flamingo fluorescent gel stain. Two-dimensional pattern of the proteins isolated from supernatant of TK173, (C) 2 h, (D) 4 h, (E) 6 h, and (F) 8 h after changing to FCS-free medium. (G) Cell secretome collected 24 h after elimination of the contaminating FCS-proteins with different washing steps. Proteins were stained with Flamingo fluorescent gel stain. Cells 2020, 9, x 3 of 19 Figure S2. 2-DE reference maps of secretomes; 150 μg proteins were loaded on an 11 cm IPG strip with a linear pH gradient PI 5–8 for IEF; 12% SDS-polyacrylamide gels were used for the second dimension. Proteins were stained with Flamingo fluorescent gel stain. Identified spots were assigned a number corresponding to that in their table. 2-DE maps from secretome of (A) TK173 control and (B) TGFβ1- treated ones. The 2-DE patterns revealed an alteration of secretome in stimulated TK173. Secretome patterns from TK173 treated with (C) ANG II and (D) PDGF. Cells 2020, 9, x 4 of 19 A Figure S3. Classification of the differentially expressed proteins upon ANG II, TGFβ1, or PDGF treatment in TK173. (A) Bar charts of the cellular component analyzed by STRAP biological function analysis in which the identified proteins from all treatments in both cell types are involved. -

The Structural Basis for Pyrophosphatase Catalysis
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector Research Article 1491 The structural basis for pyrophosphatase catalysis Pirkko Heikinheimo1,2, Jukka Lehtonen1,2, Alexander Baykov3, Reijo Lahti2, Barry S Cooperman4* and Adrian Goldman1,2* Background: Soluble inorganic pyrophosphatase (PPase), an essential Addresses: 1Turku Centre for Biotechnology, PO enzyme central to phosphorus metabolism, catalyzes the hydrolysis of the phos- Box 123, FIN-20521 Turku, Finland, 2Department phoanhydride bond in inorganic pyrophosphate. Catalysis requires divalent of Biochemistry, University of Turku, FIN-20014 Turku, Finland, 3A.N. Belozersky Institute of metal ions which affect the apparent pKas of the essential general acid and Physico-Chemical Biology, Moscow State base on the enzyme, and the pKa of the substrate. Three to five metal ions are University, Moscow 119899, Russia and required for maximal activity, depending on pH and enzyme source. A detailed 4Department of Chemistry, University of understanding of catalysis would aid both in understanding the nature of biolog- Pennsylvania, Philadelphia, PA 19104, USA. ical mechanisms of phosphoryl transfer, and in understanding the role of diva- *Corresponding authors. lent cations. Without a high-resolution complex structure such a model has E-mail: [email protected] previously been unobtainable. E-mail: [email protected] Key words: mechanism, phosphoanhydride Results: We report the first two high-resolution structures of yeast PPase, at hydrolysis, phosphoryl transfer, pyrophosphatase, 2.2 and 2.0 Å resolution with R factors of around 17 %. One structure contains refinement, structure the two activating metal ions; the other, the product (MnPi)2 as well. -

Rubisco Biogenesis and Assembly in Chlamydomonas Reinhardtii Wojciech Wietrzynski
Rubisco biogenesis and assembly in Chlamydomonas reinhardtii Wojciech Wietrzynski To cite this version: Wojciech Wietrzynski. Rubisco biogenesis and assembly in Chlamydomonas reinhardtii. Molecular biology. Université Pierre et Marie Curie - Paris VI, 2017. English. NNT : 2017PA066336. tel- 01770412 HAL Id: tel-01770412 https://tel.archives-ouvertes.fr/tel-01770412 Submitted on 19 Apr 2018 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. PhD Thesis of the Pierre and Marie Curie University (UPMC) Prepared in the Laboratory of Molecular and Membrane Physiology of the Chloroplast, UMR7141, CNRS/UPMC Doctoral school: Life Science Complexity, ED515 Presented by Wojciech Wietrzynski for the grade of Doctor of the Pierre and Marie Curie University Rubisco biogenesis and assembly in Chlamydomonas reinhardtii Defended on the 17th of October 2017 at the Institut of Physico-Chemical Biology in Paris, France Phd Jury: Angela Falciatore, CNRS/UPMC, president Michel Goldschmidt-Clermont, Univeristy of Geneva, reviewer Michael Schroda, University of Kaiserslautern, -

Protein Family Classification Using Shallow and Deep Networks ABSTRACT 1 INTRODUCTION
DeepProteomics: Protein family classification using Shallow and Deep Networks Anu Vazhayil, Vinayakumar R and Soman KP Center for Computational Engineering and Networking (CEN),Amrita School of Engineering, Coimbatore, Amrita Vishwa Vidyapeetham, India Email: [email protected], [email protected], kp [email protected] ABSTRACT The knowledge regarding the function of proteins is necessary as it gives a clear picture of biological processes. Nevertheless, there are many protein sequences found and added to the databases but lacks functional annotation. The laboratory experiments take a considerable amount of time for annotation of the sequences. This arises the need to use computational techniques to classify proteins based on their functions. In our work, we have collected the data from Swiss-Prot containing 40433 proteins which is grouped into 30 families. We pass it to recurrent neural network(RNN), long short term memory(LSTM) and gated recurrent unit(GRU) model and compare it by applying trigram with deep neural network and shallow neural network on the same dataset. Through this approach, we could achieve maximum of around 78% accuracy for the classification of protein families. Keywords Proteins, amino-acid sequences, machine learning, deep learning, recurrent neural network(RNN), long short term memory(LSTM), gated recurrent unit(GRU), deep neural networks 1 INTRODUCTION Proteins are considered to be essentials of life because it performs a variety of functions to sustain life. It performs DNA replication, transportation of molecules from one cell to another cell, accelerates metabolic reactions and several other important functions carried out within an organism. Proteins carry out these functions as specified by the informations encoded in the genes. -

An Aminoacyl-Trna Synthetase Paralog with a Catalytic Role in Histidine Biosynthesis
Proc. Natl. Acad. Sci. USA Vol. 96, pp. 8985–8990, August 1999 Biochemistry An aminoacyl-tRNA synthetase paralog with a catalytic role in histidine biosynthesis MARIE SISSLER*†,CHRISTINE DELORME†‡,JEFF BOND§,S.DUSKO EHRLICH‡,PIERRE RENAULT‡, AND CHRISTOPHER FRANCKLYN*§¶ *Department of Biochemistry, College of Medicine, Given Building, University of Vermont, Burlington, VT 05405; ‡Laboratoire de Ge´ne´tique Microbienne, INRA-CRJ, 78352 Jouy-en-Josas, France; and §Department of Microbiology and Molecular Genetics, Vermont Cancer Center, University of Vermont, Burlington, VT 05405 Edited by Paul R. Schimmel, The Scripps Research Institute, La Jolla, CA, and approved June 14, 1999 (received for review April 7, 1999) ABSTRACT In addition to their essential catalytic role in The further involvement of aaRS or aaRS-like proteins in protein biosynthesis, aminoacyl-tRNA synthetases participate amino acid biosynthesis is also suggested by the existence of in numerous other functions, including regulation of gene proteins that are based on the catalytic domains of an aaRS yet expression and amino acid biosynthesis via transamidation do not catalyze the aminoacylation reaction. A striking illus- pathways. Herein, we describe a class of aminoacyl-tRNA tration is the asparagine synthetase A (AsnA), whose recently synthetase-like (HisZ) proteins based on the catalytic core of solved structure contains a class II aaRS catalytic domain the contemporary class II histidyl-tRNA synthetase whose (closely related to AspRS and AsnRS). The role of AsnA is to members lack aminoacylation activity but are instead essen- convert aspartate to asparagine via an amidation reaction tial components of the first enzyme in histidine biosynthesis involving a transient aspartyl-adenylate (10).