Epigenetic Alterations in Skin Homing CD4+CLA+ T Cells of Atopic
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

TOX2 Is Differentially Expressed in Non-Small Cell Lung Cancer-PDF 092220
1 Tox2 is differentially expressed in non-small cell lung cancer and associates with patient 2 survival. 3 Shahan Mamoor1 4 [email protected] East Islip, NY USA 5 6 Non-small cell lung cancer (NSCLC) is the leading cause of cancer death in the United States1. 7 We mined published microarray data2,3,4 to identify differentially expressed genes in NSCLC. 8 We found that the gene encoding the thymyocyte selection-associated high mobility box protein 9 family member 2 transcription factor Tox2 was among the genes whose expression was most quantitatively different in tumors from patients with NSCLC as compared to the lung. Tox2 10 expression was significantly decreased in NSCLC tumors as compared to the lung, and lower 11 expression of Tox2 in patient tumors was significantly associated with worse overall survival. Tox2 may be important for initiation or progression of non-small cell lung cancer in humans. 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 Keywords: Tox2, NSCLC, non-small cell lung cancer, systems biology of NSCLC, targeted 27 therapeutics in NSCLC. 28 1 OF 16 1 In 2016, lung cancer resulted in the death of 158,000 Americans; 81% of all patients 2 diagnosed with lung cancer will expire within 5 years5. Non-small cell lung cancer (NSCLC) is 3 4 the most common type of lung cancer, diagnosed in 84% of patients with lung cancer, and 76% 5 of all patients with NSCLC will expire within 5 years5. The rational development of targeted 6 therapeutics to treat patients with NSCLC can be supported by an enhanced understanding of 7 8 fundamental transcriptional features of NSCLC tumors. -

Dissecting the Genetic Relationship Between Cardiovascular Risk Factors and Alzheimer's Disease
UC San Diego UC San Diego Previously Published Works Title Dissecting the genetic relationship between cardiovascular risk factors and Alzheimer's disease. Permalink https://escholarship.org/uc/item/7137q6g1 Journal Acta neuropathologica, 137(2) ISSN 0001-6322 Authors Broce, Iris J Tan, Chin Hong Fan, Chun Chieh et al. Publication Date 2019-02-01 DOI 10.1007/s00401-018-1928-6 Peer reviewed eScholarship.org Powered by the California Digital Library University of California Acta Neuropathologica https://doi.org/10.1007/s00401-018-1928-6 ORIGINAL PAPER Dissecting the genetic relationship between cardiovascular risk factors and Alzheimer’s disease Iris J. Broce1 · Chin Hong Tan1,2 · Chun Chieh Fan3 · Iris Jansen4 · Jeanne E. Savage4 · Aree Witoelar5 · Natalie Wen6 · Christopher P. Hess1 · William P. Dillon1 · Christine M. Glastonbury1 · Maria Glymour7 · Jennifer S. Yokoyama8 · Fanny M. Elahi8 · Gil D. Rabinovici8 · Bruce L. Miller8 · Elizabeth C. Mormino9 · Reisa A. Sperling10,11 · David A. Bennett12 · Linda K. McEvoy13 · James B. Brewer13,14,15 · Howard H. Feldman14 · Bradley T. Hyman10 · Margaret Pericak‑Vance16 · Jonathan L. Haines17,18 · Lindsay A. Farrer19,20,21,22,23 · Richard Mayeux24,25,26 · Gerard D. Schellenberg27 · Kristine Yafe7,8,28 · Leo P. Sugrue1 · Anders M. Dale3,13,14 · Danielle Posthuma4 · Ole A. Andreassen5 · Celeste M. Karch6 · Rahul S. Desikan1 Received: 22 September 2018 / Revised: 28 October 2018 / Accepted: 28 October 2018 © Springer-Verlag GmbH Germany, part of Springer Nature 2018 Abstract Cardiovascular (CV)- and lifestyle-associated risk factors (RFs) are increasingly recognized as important for Alzheimer’s disease (AD) pathogenesis. Beyond the ε4 allele of apolipoprotein E (APOE), comparatively little is known about whether CV-associated genes also increase risk for AD. -

TOX As a Potential Target for Immunotherapy in Lymphocytic Malignancies Chaofeng Liang1,2, Shuxin Huang1, Yujie Zhao1, Shaohua Chen1* and Yangqiu Li1*
Liang et al. Biomarker Research (2021) 9:20 https://doi.org/10.1186/s40364-021-00275-y REVIEW Open Access TOX as a potential target for immunotherapy in lymphocytic malignancies Chaofeng Liang1,2, Shuxin Huang1, Yujie Zhao1, Shaohua Chen1* and Yangqiu Li1* Abstract TOX (thymocyte selection-associated HMG BOX) is a member of a family of transcriptional factors that contain the highly conserved high mobility group box (HMG-box) region. Increasing studies have shown that TOX is involved in maintaining tumors and promoting T cell exhaustion. In this review, we summarized the biological functions of TOX and its contribution as related to lymphocytic malignancies. We also discussed the potential role of TOX as an immune biomarker and target in immunotherapy for hematological malignancies. Keywords: Thymocyte selection-associated HMG BOX, Biological function, T cell exhaustion, Lymphocytic malignancies, Immune biomarker, Immunotherapy Background located on different chromosomes and have different TOX (thymocyte selection-associated HMG BOX), a functions (Fig. 1, Table 1)[16]. transcription factor, belongs to the high mobility group box (HMG-box) superfamily. In 2002, Wilkinson et al. TOX demonstrated for the first time that TOX plays an im- TOX, also known as KIAA0808, is located on q12.1 of portant role in the differentiation of CD4 + CD8+ human chromosome 8 and has one transcript. The double-positive thymocytes by gene microarray technol- length of the TOX mRNA is 4076 bp, and it contains ogy. Numerous studies have shown that TOX plays a nine exons that translate into a protein consisting of 526 vital role in the development and formation of various amino acids. -

A Clinicopathological and Molecular Genetic Analysis of Low-Grade Glioma in Adults
A CLINICOPATHOLOGICAL AND MOLECULAR GENETIC ANALYSIS OF LOW-GRADE GLIOMA IN ADULTS Presented by ANUSHREE SINGH MSc A thesis submitted in partial fulfilment of the requirements of the University of Wolverhampton for the degree of Doctor of Philosophy Brain Tumour Research Centre Research Institute in Healthcare Sciences Faculty of Science and Engineering University of Wolverhampton November 2014 i DECLARATION This work or any part thereof has not previously been presented in any form to the University or to any other body whether for the purposes of assessment, publication or for any other purpose (unless otherwise indicated). Save for any express acknowledgments, references and/or bibliographies cited in the work, I confirm that the intellectual content of the work is the result of my own efforts and of no other person. The right of Anushree Singh to be identified as author of this work is asserted in accordance with ss.77 and 78 of the Copyright, Designs and Patents Act 1988. At this date copyright is owned by the author. Signature: Anushree Date: 30th November 2014 ii ABSTRACT The aim of the study was to identify molecular markers that can determine progression of low grade glioma. This was done using various approaches such as IDH1 and IDH2 mutation analysis, MGMT methylation analysis, copy number analysis using array comparative genomic hybridisation and identification of differentially expressed miRNAs using miRNA microarray analysis. IDH1 mutation was present at a frequency of 71% in low grade glioma and was identified as an independent marker for improved OS in a multivariate analysis, which confirms the previous findings in low grade glioma studies. -

Transcriptional Control of Tissue-Resident Memory T Cell Generation
Transcriptional control of tissue-resident memory T cell generation Filip Cvetkovski Submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Graduate School of Arts and Sciences COLUMBIA UNIVERSITY 2019 © 2019 Filip Cvetkovski All rights reserved ABSTRACT Transcriptional control of tissue-resident memory T cell generation Filip Cvetkovski Tissue-resident memory T cells (TRM) are a non-circulating subset of memory that are maintained at sites of pathogen entry and mediate optimal protection against reinfection. Lung TRM can be generated in response to respiratory infection or vaccination, however, the molecular pathways involved in CD4+TRM establishment have not been defined. Here, we performed transcriptional profiling of influenza-specific lung CD4+TRM following influenza infection to identify pathways implicated in CD4+TRM generation and homeostasis. Lung CD4+TRM displayed a unique transcriptional profile distinct from spleen memory, including up-regulation of a gene network induced by the transcription factor IRF4, a known regulator of effector T cell differentiation. In addition, the gene expression profile of lung CD4+TRM was enriched in gene sets previously described in tissue-resident regulatory T cells. Up-regulation of immunomodulatory molecules such as CTLA-4, PD-1, and ICOS, suggested a potential regulatory role for CD4+TRM in tissues. Using loss-of-function genetic experiments in mice, we demonstrate that IRF4 is required for the generation of lung-localized pathogen-specific effector CD4+T cells during acute influenza infection. Influenza-specific IRF4−/− T cells failed to fully express CD44, and maintained high levels of CD62L compared to wild type, suggesting a defect in complete differentiation into lung-tropic effector T cells. -

Targeting Hedgehog Signalling Through the Ubiquitylation Process: the Multiple Roles of the HECT-E3 Ligase Itch
cells Review Targeting Hedgehog Signalling through the Ubiquitylation Process: The Multiple Roles of the HECT-E3 Ligase Itch Paola Infante 1,†, Ludovica Lospinoso Severini 2,† , Flavia Bernardi 2, Francesca Bufalieri 2 and Lucia Di Marcotullio 2,3,* 1 Center for Life NanoScience@Sapienza, Istituto Italiano di Tecnologia, 00161 Rome, Italy; [email protected] 2 Department of Molecular Medicine, University of Rome La Sapienza, 00161 Rome, Italy; [email protected] (L.L.S.); fl[email protected] (F.B.); [email protected] (F.B.) 3 Istituto Pasteur-Fondazione Cenci Bolognetti, University of Rome La Sapienza, 00161 Rome, Italy * Correspondence: [email protected]; Tel.: +39-06-49255657 † These authors contributed equally to this work. Received: 28 December 2018; Accepted: 26 January 2019; Published: 29 January 2019 Abstract: Hedgehog signalling (Hh) is a developmental conserved pathway strongly involved in cancers when deregulated. This important pathway is orchestrated by numerous regulators, transduces through distinct routes and is finely tuned at multiple levels. In this regard, ubiquitylation processes stand as essential for controlling Hh pathway output. Although this post-translational modification governs proteins turnover, it is also implicated in non-proteolytic events, thereby regulating the most important cellular functions. The HECT E3 ligase Itch, well known to control immune response, is emerging to have a pivotal role in tumorigenesis. By illustrating Itch specificities on Hh signalling key components, here we review the role of this HECT E3 ubiquitin ligase in suppressing Hh-dependent tumours and explore its potential as promising target for innovative therapeutic approaches. Keywords: Hedgehog; cancer; ubiquitylation; Itch; Numb; β-arrestin2; GLI1; SuFu; Patched1 1. -

Supplementary Table S5. Differentially Expressed Gene Lists of PD-1High CD39+ CD8 Tils According to 4-1BB Expression Compared to PD-1+ CD39- CD8 Tils
BMJ Publishing Group Limited (BMJ) disclaims all liability and responsibility arising from any reliance Supplemental material placed on this supplemental material which has been supplied by the author(s) J Immunother Cancer Supplementary Table S5. Differentially expressed gene lists of PD-1high CD39+ CD8 TILs according to 4-1BB expression compared to PD-1+ CD39- CD8 TILs Up- or down- regulated genes in Up- or down- regulated genes Up- or down- regulated genes only PD-1high CD39+ CD8 TILs only in 4-1BBneg PD-1high CD39+ in 4-1BBpos PD-1high CD39+ CD8 compared to PD-1+ CD39- CD8 CD8 TILs compared to PD-1+ TILs compared to PD-1+ CD39- TILs CD39- CD8 TILs CD8 TILs IL7R KLRG1 TNFSF4 ENTPD1 DHRS3 LEF1 ITGA5 MKI67 PZP KLF3 RYR2 SIK1B ANK3 LYST PPP1R3B ETV1 ADAM28 H2AC13 CCR7 GFOD1 RASGRP2 ITGAX MAST4 RAD51AP1 MYO1E CLCF1 NEBL S1PR5 VCL MPP7 MS4A6A PHLDB1 GFPT2 TNF RPL3 SPRY4 VCAM1 B4GALT5 TIPARP TNS3 PDCD1 POLQ AKAP5 IL6ST LY9 PLXND1 PLEKHA1 NEU1 DGKH SPRY2 PLEKHG3 IKZF4 MTX3 PARK7 ATP8B4 SYT11 PTGER4 SORL1 RAB11FIP5 BRCA1 MAP4K3 NCR1 CCR4 S1PR1 PDE8A IFIT2 EPHA4 ARHGEF12 PAICS PELI2 LAT2 GPRASP1 TTN RPLP0 IL4I1 AUTS2 RPS3 CDCA3 NHS LONRF2 CDC42EP3 SLCO3A1 RRM2 ADAMTSL4 INPP5F ARHGAP31 ESCO2 ADRB2 CSF1 WDHD1 GOLIM4 CDK5RAP1 CD69 GLUL HJURP SHC4 GNLY TTC9 HELLS DPP4 IL23A PITPNC1 TOX ARHGEF9 EXO1 SLC4A4 CKAP4 CARMIL3 NHSL2 DZIP3 GINS1 FUT8 UBASH3B CDCA5 PDE7B SOGA1 CDC45 NR3C2 TRIB1 KIF14 TRAF5 LIMS1 PPP1R2C TNFRSF9 KLRC2 POLA1 CD80 ATP10D CDCA8 SETD7 IER2 PATL2 CCDC141 CD84 HSPA6 CYB561 MPHOSPH9 CLSPN KLRC1 PTMS SCML4 ZBTB10 CCL3 CA5B PIP5K1B WNT9A CCNH GEM IL18RAP GGH SARDH B3GNT7 C13orf46 SBF2 IKZF3 ZMAT1 TCF7 NECTIN1 H3C7 FOS PAG1 HECA SLC4A10 SLC35G2 PER1 P2RY1 NFKBIA WDR76 PLAUR KDM1A H1-5 TSHZ2 FAM102B HMMR GPR132 CCRL2 PARP8 A2M ST8SIA1 NUF2 IL5RA RBPMS UBE2T USP53 EEF1A1 PLAC8 LGR6 TMEM123 NEK2 SNAP47 PTGIS SH2B3 P2RY8 S100PBP PLEKHA7 CLNK CRIM1 MGAT5 YBX3 TP53INP1 DTL CFH FEZ1 MYB FRMD4B TSPAN5 STIL ITGA2 GOLGA6L10 MYBL2 AHI1 CAND2 GZMB RBPJ PELI1 HSPA1B KCNK5 GOLGA6L9 TICRR TPRG1 UBE2C AURKA Leem G, et al. -

Table S1| Differential Expression Analysis of the Atopy Transcriptome
Table S1| Differential expression analysis of the atopy transcriptome in CD4+ T-cell responses to allergens in atopic and nonatopic subjects Probe ID S.test Gene Symbol Gene Description Chromosome Statistic Location 7994280 10.32 IL4R Interleukin 4 receptor 16p11.2-12.1 8143383 8.95 --- --- --- 7974689 8.50 DACT1 Dapper, antagonist of beta-catenin, homolog 1 14q23.1 8102415 7.59 CAMK2D Calcium/calmodulin-dependent protein kinase II delta 4q26 7950743 7.58 RAB30 RAB30, member RAS oncogene family 11q12-q14 8136580 7.54 RAB19B GTP-binding protein RAB19B 7q34 8043504 7.45 MAL Mal, T-cell differentiation protein 2cen-q13 8087739 7.27 CISH Cytokine inducible SH2-containing protein 3p21.3 8000413 7.17 NSMCE1 Non-SMC element 1 homolog (S. cerevisiae) 16p12.1 8021301 7.15 RAB27B RAB27B, member RAS oncogene family 18q21.2 8143367 6.83 SLC37A3 Solute carrier family 37 member 3 7q34 8152976 6.65 TMEM71 Transmembrane protein 71 8q24.22 7931914 6.56 IL2R Interleukin 2 receptor, alpha 10p15-p14 8014768 6.43 PLXDC1 Plexin domain containing 1 17q21.1 8056222 6.43 DPP4 Dipeptidyl-peptidase 4 (CD26) 2q24.3 7917697 6.40 GFI1 Growth factor independent 1 1p22 7903507 6.39 FAM102B Family with sequence similarity 102, member B 1p13.3 7968236 5.96 RASL11A RAS-like, family 11, member A --- 7912537 5.95 DHRS3 Dehydrogenase/reductase (SDR family) member 3 1p36.1 7963491 5.83 KRT1 Keratin 1 (epidermolytic hyperkeratosis) 12q12-q13 7903786 5.72 CSF1 Colony stimulating factor 1 (macrophage) 1p21-p13 8019061 5.67 SGSH N-sulfoglucosamine sulfohydrolase (sulfamidase) 17q25.3 -

TOX3 Is Expressed in Mammary ER+ Epithelial Cells and Regulates ER
Seksenyan et al. BMC Cancer (2015) 15:22 DOI 10.1186/s12885-015-1018-2 RESEARCH ARTICLE Open Access TOX3 is expressed in mammary ER+ epithelial cells and regulates ER target genes in luminal breast cancer Akop Seksenyan1, Asha Kadavallore1, Ann E Walts2, Brian de la Torre1, Dror Berel3,4, Samuel P Strom5,6, Parinaz Aliahmad1, Vincent A Funari5 and Jonathan Kaye1,3,7* Abstract Background: A breast cancer susceptibility locus has been mapped to the gene encoding TOX3. Little is known regarding the expression pattern or biological role of TOX3 in breast cancer or in the mammary gland. Here we analyzed TOX3 expression in murine and human mammary glands and in molecular subtypes of breast cancer, and assessed its ability to alter the biology of breast cancer cells. Methods: We used a cell sorting strategy, followed by quantitative real-time PCR, to study TOX3 gene expression in the mouse mammary gland. To study the expression of this nuclear protein in human mammary glands and breast tumors, we generated a rabbit monoclonal antibody specific for human TOX3. In vitro studies were performed on MCF7, BT474 and MDA-MB-231 cell lines to study the effects of TOX3 modulation on gene expression in the context of breast cancer cells. Results: We found TOX3 expression in estrogen receptor-positive mammary epithelial cells, including progenitor cells. A subset of breast tumors also highly expresses TOX3, with poor outcome associated with high expression of TOX3 in luminal B breast cancers. We also demonstrate the ability of TOX3 to alter gene expression in MCF7 luminal breast cancer cells, including cancer relevant genes TFF1 and CXCR4. -
Primepcr™Assay Validation Report
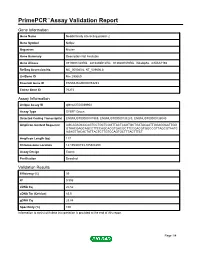
PrimePCR™Assay Validation Report Gene Information Gene Name Nedd4 family interacting protein 2 Gene Symbol Ndfip2 Organism Mouse Gene Summary Description Not Available Gene Aliases 0710001O20Rik, 2810436B12Rik, 9130207N19Rik, N4wbp5a, mKIAA1165 RefSeq Accession No. NC_000080.6, NT_039606.8 UniGene ID Mm.290669 Ensembl Gene ID ENSMUSG00000053253 Entrez Gene ID 76273 Assay Information Unique Assay ID qMmuCED0039954 Assay Type SYBR® Green Detected Coding Transcript(s) ENSMUST00000181969, ENSMUST00000138283, ENSMUST00000136040 Amplicon Context Sequence AGCAGAGCCCAGTCCTGCTCGGTTACTCAGTGCTGATACAATTGGAGGAATTGG GTAACGAGCAGCCTTCCAGCACGTGACGCTTCCGACGTGGCCGTTAGCGTAATC AGAGTTACACTATTACTCTTGTCCAGTGCTTTACTTTCT Amplicon Length (bp) 117 Chromosome Location 14:105308153-105308299 Assay Design Exonic Purification Desalted Validation Results Efficiency (%) 99 R2 0.996 cDNA Cq 20.52 cDNA Tm (Celsius) 83.5 gDNA Cq 23.84 Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 1/4 PrimePCR™Assay Validation Report Ndfip2, Mouse Amplification Plot Amplification of cDNA generated from 25 ng of universal reference RNA Melt Peak Melt curve analysis of above amplification Standard Curve Standard curve generated using 20 million copies of template diluted 10-fold to 20 copies Page 2/4 PrimePCR™Assay Validation Report Products used to generate validation data Real-Time PCR Instrument CFX384 Real-Time PCR Detection System Reverse Transcription Reagent iScript™ Advanced cDNA Synthesis Kit for RT-qPCR Real-Time PCR Supermix SsoAdvanced™ SYBR® Green Supermix Experimental Sample qPCR Mouse Reference Total RNA Data Interpretation Unique Assay ID This is a unique identifier that can be used to identify the assay in the literature and online. Detected Coding Transcript(s) This is a list of the Ensembl transcript ID(s) that this assay will detect. -

Greg's Awesome Thesis
Analysis of alignment error and sitewise constraint in mammalian comparative genomics Gregory Jordan European Bioinformatics Institute University of Cambridge A dissertation submitted for the degree of Doctor of Philosophy November 30, 2011 To my parents, who kept us thinking and playing This dissertation is the result of my own work and includes nothing which is the out- come of work done in collaboration except where specifically indicated in the text and acknowledgements. This dissertation is not substantially the same as any I have submitted for a degree, diploma or other qualification at any other university, and no part has already been, or is currently being submitted for any degree, diploma or other qualification. This dissertation does not exceed the specified length limit of 60,000 words as defined by the Biology Degree Committee. November 30, 2011 Gregory Jordan ii Analysis of alignment error and sitewise constraint in mammalian comparative genomics Summary Gregory Jordan November 30, 2011 Darwin College Insight into the evolution of protein-coding genes can be gained from the use of phylogenetic codon models. Recently sequenced mammalian genomes and powerful analysis methods developed over the past decade provide the potential to globally measure the impact of natural selection on pro- tein sequences at a fine scale. The detection of positive selection in particular is of great interest, with relevance to the study of host-parasite conflicts, immune system evolution and adaptive dif- ferences between species. This thesis examines the performance of methods for detecting positive selection first with a series of simulation experiments, and then with two empirical studies in mammals and primates. -

TOX3 Is a Favorable Prognostic Indicator and Potential Immunomodulatory Factor in Lung Adenocarcinoma
4144 ONCOLOGY LETTERS 18: 4144-4152, 2019 TOX3 is a favorable prognostic indicator and potential immunomodulatory factor in lung adenocarcinoma DE ZENG1, HAOYU LIN2, JIANXIONG CUI2 and WEIQUAN LIANG2 1Department of Medical Oncology, Cancer Hospital of Shantou University Medical College; 2Department of Breast and Thyroid Surgery, The First Affiliated Hospital of Shantou University Medical College, Shantou, Guangdong 515000, P.R. China Received February 13, 2018; Accepted January 14, 2019 DOI: 10.3892/ol.2019.10748 Abstract. Thymocyte selection-associated high mobility and cytotoxic T lymphocyte-associated protein 4 (CTLA4), as group box (TOX) genes represent a novel family of genes. well as the development of novel immunotherapy agents, lung Deregulated expression of TOXs has been reported in a cancer treatments have improved notably in terms of disease variety of cancer types, including lung cancer. It has also control and survival outcomes (3). However, the success rate been reported that TOXs are crucial regulators of the immune of immunotherapy remains low, and a large number of cancer system. The present study systematically evaluated the prog- types, including lung cancer, remain largely insensitive or nostic values of TOX family members using a set of publicly refractory to PD-1, PD-L1 and CTLA4 blockade (2,4-6). accessible databases, including Oncomine, Kaplan-Meier Therefore, research has focused on identifying novel immu- plotter and cBioPortal. It was revealed that TOX expression notherapy targets or immunomodulatory factors that can profiles differed between lung cancer and normal tissues, be exploited to enhance the response to current immuno- and high expression of TOX mRNAs generally predicted therapeutic agents and address immunotherapy resistance in improved survival outcomes.