Antibiotics from Deep-Sea Microorganisms: Current Discoveries and Perspectives
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Chapter 10: Classification of Microorganisms
Chapter 10: Classification of Microorganisms 1. The Taxonomic Hierarchy 2. Methods of Identification 1. The Taxonomic Hierarchy Phylogenetic Tree of the 3 Domains Taxonomic Hierarchy • 8 successive taxa are used to classify each species: Domain Kingdom Phylum Class Order Family Genus **species can also contain different strains** Species Scientific Nomenclature To avoid confusion, every type of organism must be referred to in a consistent way. The current system of nomenclature (naming) has been in use since the 18th century: • every type of organism is referred by its genus name followed by its specific epithet (i.e., species name) Homo sapiens (H. sapiens) Escherichia coli (E. coli) • name should be in italics and only the genus is capitalized which can also be abbreviated • names are Latin (or “Latinized” Greek) with the genus being a noun and the specific epithet an adjective **strain info can be listed after the specific epithet (e.g., E. coli DH5α)** 2. Methods of Identification Biochemical Testing In addition to morphological (i.e., appearance under the microscope) and differential staining characteristics, microorganisms can also be identified by their biochemical “signatures”: • the nutrient requirements and metabolic “by-products” of of a particular microorganism • different growth media can be used to test the physiological characteristics of a microorganism • e.g., medium with lactose only as energy source • e.g., medium that reveals H2S production **appearance on test medium reveals + or – result!** Commercial devices for rapid Identification Perform multiple tests simultaneously Enterotube II Such devices involve the simultaneous inoculation of various test media: • ~24 hrs later the panel of results reveals ID of organism! Use of Dichotomous Keys Series of “yes/no” biochemical tests to ID organism. -

Anoxygenic Photosynthesis in Photolithotrophic Sulfur Bacteria and Their Role in Detoxication of Hydrogen Sulfide
antioxidants Review Anoxygenic Photosynthesis in Photolithotrophic Sulfur Bacteria and Their Role in Detoxication of Hydrogen Sulfide Ivan Kushkevych 1,* , Veronika Bosáková 1,2 , Monika Vítˇezová 1 and Simon K.-M. R. Rittmann 3,* 1 Department of Experimental Biology, Faculty of Science, Masaryk University, 62500 Brno, Czech Republic; [email protected] (V.B.); [email protected] (M.V.) 2 Department of Biology, Faculty of Medicine, Masaryk University, 62500 Brno, Czech Republic 3 Archaea Physiology & Biotechnology Group, Department of Functional and Evolutionary Ecology, Universität Wien, 1090 Vienna, Austria * Correspondence: [email protected] (I.K.); [email protected] (S.K.-M.R.R.); Tel.: +420-549-495-315 (I.K.); +431-427-776-513 (S.K.-M.R.R.) Abstract: Hydrogen sulfide is a toxic compound that can affect various groups of water microorgan- isms. Photolithotrophic sulfur bacteria including Chromatiaceae and Chlorobiaceae are able to convert inorganic substrate (hydrogen sulfide and carbon dioxide) into organic matter deriving energy from photosynthesis. This process takes place in the absence of molecular oxygen and is referred to as anoxygenic photosynthesis, in which exogenous electron donors are needed. These donors may be reduced sulfur compounds such as hydrogen sulfide. This paper deals with the description of this metabolic process, representatives of the above-mentioned families, and discusses the possibility using anoxygenic phototrophic microorganisms for the detoxification of toxic hydrogen sulfide. Moreover, their general characteristics, morphology, metabolism, and taxonomy are described as Citation: Kushkevych, I.; Bosáková, well as the conditions for isolation and cultivation of these microorganisms will be presented. V.; Vítˇezová,M.; Rittmann, S.K.-M.R. -

Revised Glossary for AQA GCSE Biology Student Book
Biology Glossary amino acids small molecules from which proteins are A built abiotic factor physical or non-living conditions amylase a digestive enzyme (carbohydrase) that that affect the distribution of a population in an breaks down starch ecosystem, such as light, temperature, soil pH anaerobic respiration respiration without using absorption the process by which soluble products oxygen of digestion move into the blood from the small intestine antibacterial chemicals chemicals produced by plants as a defence mechanism; the amount abstinence method of contraception whereby the produced will increase if the plant is under attack couple refrains from intercourse, particularly when an egg might be in the oviduct antibiotic e.g. penicillin; medicines that work inside the body to kill bacterial pathogens accommodation ability of the eyes to change focus antibody protein normally present in the body acid rain rain water which is made more acidic by or produced in response to an antigen, which it pollutant gases neutralises, thus producing an immune response active site the place on an enzyme where the antimicrobial resistance (AMR) an increasing substrate molecule binds problem in the twenty-first century whereby active transport in active transport, cells use energy bacteria have evolved to develop resistance against to transport substances through cell membranes antibiotics due to their overuse against a concentration gradient antiretroviral drugs drugs used to treat HIV adaptation features that organisms have to help infections; they -

Marine Microorganisms: Evolution and Solution to Pollution Fu L Li1, Wang B1,2
COMMENTARY Marine microorganisms: Evolution and solution to pollution Fu L Li1, Wang B1,2 Li FL, Wang B. Marine microorganisms: Evolution and solution to pollution. J Mar Microbiol. 2018;2(1):4-5. nce ocean nurtured life, now she needs our care. Marine microorganism will be an opportunity to further understand ourselves and to seek for new Ois the host of ocean in all ages. We should learn from them humbly. methods of fighting old infections. Marine microorganism is tightly bond with human during the history of evolution and nowadays’ environment pollution. Along with industrial revolution, our marine ecosystem suffered serious pollutions. Microplastics are tiny plastic particles (<5 mm) (Figure 1B), Although the topic is still in debate, life is probably originated from which poison marine lives. Because these microplastics are very hard to be submarine in hydrothermal vent systems (1). In the journey of evolution, our degraded, it is predicted that there will be more microplastics than fish in biosphere was completely dominated by microbes for a very long time (Figure ocean by the year 2050 (7). Since marine sediments are considered as the sink 1A). Human being evolves with those microorganisms. Consequently, of microplastics and marine microbes are key dwellers of marine sediments, the influences of microorganisms can be found in all aspects of human more attention should be paid on the interactions between microplastics biology. More than 65% of our genes originated with bacteria, archaea, and and marine microbes. Actually, a call for this has been published in 2011 unicellular eukaryotes, including those genes responsible for host-microbe (8). -

Phytoplankton As Key Mediators of the Biological Carbon Pump: Their Responses to a Changing Climate
sustainability Review Phytoplankton as Key Mediators of the Biological Carbon Pump: Their Responses to a Changing Climate Samarpita Basu * ID and Katherine R. M. Mackey Earth System Science, University of California Irvine, Irvine, CA 92697, USA; [email protected] * Correspondence: [email protected] Received: 7 January 2018; Accepted: 12 March 2018; Published: 19 March 2018 Abstract: The world’s oceans are a major sink for atmospheric carbon dioxide (CO2). The biological carbon pump plays a vital role in the net transfer of CO2 from the atmosphere to the oceans and then to the sediments, subsequently maintaining atmospheric CO2 at significantly lower levels than would be the case if it did not exist. The efficiency of the biological pump is a function of phytoplankton physiology and community structure, which are in turn governed by the physical and chemical conditions of the ocean. However, only a few studies have focused on the importance of phytoplankton community structure to the biological pump. Because global change is expected to influence carbon and nutrient availability, temperature and light (via stratification), an improved understanding of how phytoplankton community size structure will respond in the future is required to gain insight into the biological pump and the ability of the ocean to act as a long-term sink for atmospheric CO2. This review article aims to explore the potential impacts of predicted changes in global temperature and the carbonate system on phytoplankton cell size, species and elemental composition, so as to shed light on the ability of the biological pump to sequester carbon in the future ocean. -

THE CASE AGAINST Marine Mammals in Captivity Authors: Naomi A
s l a m m a y t T i M S N v I i A e G t A n i p E S r a A C a C E H n T M i THE CASE AGAINST Marine Mammals in Captivity The Humane Society of the United State s/ World Society for the Protection of Animals 2009 1 1 1 2 0 A M , n o t s o g B r o . 1 a 0 s 2 u - e a t i p s u S w , t e e r t S h t u o S 9 8 THE CASE AGAINST Marine Mammals in Captivity Authors: Naomi A. Rose, E.C.M. Parsons, and Richard Farinato, 4th edition Editors: Naomi A. Rose and Debra Firmani, 4th edition ©2009 The Humane Society of the United States and the World Society for the Protection of Animals. All rights reserved. ©2008 The HSUS. All rights reserved. Printed on recycled paper, acid free and elemental chlorine free, with soy-based ink. Cover: ©iStockphoto.com/Ying Ying Wong Overview n the debate over marine mammals in captivity, the of the natural environment. The truth is that marine mammals have evolved physically and behaviorally to survive these rigors. public display industry maintains that marine mammal For example, nearly every kind of marine mammal, from sea lion Iexhibits serve a valuable conservation function, people to dolphin, travels large distances daily in a search for food. In learn important information from seeing live animals, and captivity, natural feeding and foraging patterns are completely lost. -
Lokiarchaeota: Biologists Discover 'Missing Link' Microorganism
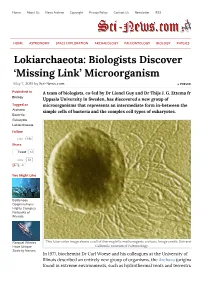
Home About Us News Archive Copyright Privacy Policy Contact Us Newsletter RSS HOME ASTRONOMY SPACE EXPLORATION ARCHAEOLOGY PALEONTOLOGY BIOLOGY PHYSICS Lokiarchaeota: Biologists Discover ‘Missing Link’ Microorganism May 7, 2015 by Sci-News.com « PREVIOUS Published in A team of biologists, co-led by Dr Lionel Guy and Dr Thijs J. G. Ettema from Biology Uppsala University in Sweden, has discovered a new group of Tagged as microorganisms that represents an intermediate form in-between the Archaea simple cells of bacteria and the complex cell types of eukaryotes. Bacteria Eukaryote Lokiarchaeota Follow Like 16k Share Tweet 12 Like 58 41 You Might Like Bottlenose Dolphins Form Highly Complex Networks of Friends Rorqual Whales This false-color image shows a cell of thermophilic methanogenic archaea. Image credit: University of Have Unique California Museum of Paleontology. Stretchy Nerves In 1977, biochemist Dr Carl Woese and his colleagues at the University of Illinois described an entirely new group of organisms, the Archaea (originally found in extreme environments, such as hydrothermal vents and terrestrial hot springs). The scientists were studying relationships among the prokaryotes using DNA Extinction of sequences, and found that Archaea have distinct molecular characteristics World’s Largest separating them from bacteria as well as from eukaryotes. They proposed that Herbivores May Lead to Empty life can be divided into three domains: Eukaryota, Eubacteria, and Landscapes, Say Archaebacteria. Researchers Despite that archaeal cells were simple and small like bacteria, scientists found that Archaea were more closely related to organisms with complex cell types, a group collectively known as ‘eukaryotes.’ This observation has puzzled Sichuan Bush biologists for years. -

Marine Microplankton Ecology Reading
Marine Microplankton Ecology Reading Microbes dominate our planet, especially the Earth’s oceans. The distinguishing feature of microorganisms is their small size, usually defined as less than 200 micrometers (µm); they are all invisible to the naked eye. As a group, sea microbes are extremely diverse, and extremely versatile with respect to their abilities to make and eat food. All marine microbes are too small to swim against the current and are therefore classified as plankton. First we will discuss several ways to classify marine microbes. 1. Size Planktonic marine organisms can be divided into the following size categories: Category Size femtoplankton <0.2 µm picoplankton 0.2-2 µm nanoplankton 2-20 µm microplankton 20-200 µm mesoplankton 200-2000 µm In this laboratory we are concerned with the microscopic portion of the plankton, less than 200 µm. These organisms are not visible to the naked eye (Figure 1). Figure 1. Size classes of marine plankton 2. Type A. Viruses Viruses are the smallest and simplest microplankton. They range from 0.01 to 0.3 um in diameter. Externally, viruses have a capsid, or protein coat. Viruses can also have simple or complex external morphologies with tail fibers and structures that are used to inject DNA or RNA into their host. Viruses have little internal morphology. They do not have a nucleus or organelles. They do not have chlorophyll. Inside a virus there is only nucleic acid, either DNA or RNA. Viruses do not grow and have no metabolism. Marine viruses are highly abundant. There are up to 10 billion in one liter of seawater! B. -

Laboratory Exercises in Microbiology: Discovering the Unseen World Through Hands-On Investigation
City University of New York (CUNY) CUNY Academic Works Open Educational Resources Queensborough Community College 2016 Laboratory Exercises in Microbiology: Discovering the Unseen World Through Hands-On Investigation Joan Petersen CUNY Queensborough Community College Susan McLaughlin CUNY Queensborough Community College How does access to this work benefit ou?y Let us know! More information about this work at: https://academicworks.cuny.edu/qb_oers/16 Discover additional works at: https://academicworks.cuny.edu This work is made publicly available by the City University of New York (CUNY). Contact: [email protected] Laboratory Exercises in Microbiology: Discovering the Unseen World through Hands-On Investigation By Dr. Susan McLaughlin & Dr. Joan Petersen Queensborough Community College Laboratory Exercises in Microbiology: Discovering the Unseen World through Hands-On Investigation Table of Contents Preface………………………………………………………………………………………i Acknowledgments…………………………………………………………………………..ii Microbiology Lab Safety Instructions…………………………………………………...... iii Lab 1. Introduction to Microscopy and Diversity of Cell Types……………………......... 1 Lab 2. Introduction to Aseptic Techniques and Growth Media………………………...... 19 Lab 3. Preparation of Bacterial Smears and Introduction to Staining…………………...... 37 Lab 4. Acid fast and Endospore Staining……………………………………………......... 49 Lab 5. Metabolic Activities of Bacteria…………………………………………….…....... 59 Lab 6. Dichotomous Keys……………………………………………………………......... 77 Lab 7. The Effect of Physical Factors on Microbial Growth……………………………... 85 Lab 8. Chemical Control of Microbial Growth—Disinfectants and Antibiotics…………. 99 Lab 9. The Microbiology of Milk and Food………………………………………………. 111 Lab 10. The Eukaryotes………………………………………………………………........ 123 Lab 11. Clinical Microbiology I; Anaerobic pathogens; Vectors of Infectious Disease….. 141 Lab 12. Clinical Microbiology II—Immunology and the Biolog System………………… 153 Lab 13. Putting it all Together: Case Studies in Microbiology…………………………… 163 Appendix I. -

Human Microbiome: Your Body Is an Ecosystem
Human Microbiome: Your Body Is an Ecosystem This StepRead is based on an article provided by the American Museum of Natural History. What Is an Ecosystem? An ecosystem is a community of living things. The living things in an ecosystem interact with each other and with the non-living things around them. One example of an ecosystem is a forest. Every forest has a mix of living things, like plants and animals, and non-living things, like air, sunlight, rocks, and water. The mix of living and non-living things in each forest is unique. It is different from the mix of living and non-living things in any other ecosystem. You Are an Ecosystem The human body is also an ecosystem. There are trillions tiny organisms living in and on it. These organisms are known as microbes and include bacteria, viruses, and fungi. There are more of them living on just your skin right now than there are people on Earth. And there are a thousand times more than that in your gut! All the microbes in and on the human body form communities. The human body is an ecosystem. It is home to trillions of microbes. These communities are part of the ecosystem of the human Photo Credit: Gaby D’Alessandro/AMNH body. Together, all of these communities are known as the human microbiome. No two human microbiomes are the same. Because of this, you are a unique ecosystem. There is no other ecosystem like your body. Humans & Microbes Microbes have been around for more than 3.5 billion years. -

Marine Extremophiles: a Source of Hydrolases for Biotechnological Applications
Mar. Drugs 2015, 13, 1925-1965; doi:10.3390/md13041925 OPEN ACCESS marine drugs ISSN 1660-3397 www.mdpi.com/journal/marinedrugs Article Marine Extremophiles: A Source of Hydrolases for Biotechnological Applications Gabriel Zamith Leal Dalmaso 1,2, Davis Ferreira 3 and Alane Beatriz Vermelho 1,* 1 BIOINOVAR—Biotechnology laboratories: Biocatalysis, Bioproducts and Bioenergy, Institute of Microbiology Paulo de Góes, Federal University of Rio de Janeiro, Av. Carlos Chagas Filho, 373, 21941-902 Rio de Janeiro, Brazil; E-Mail: [email protected] 2 Graduate Program in Plant Biotechnology, Health and Science Centre, Federal University of Rio de Janeiro, Av. Carlos Chagas Filho, 373, 21941-902 Rio de Janeiro, Brazil 3 BIOINOVAR—Biotechnology Laboratories: Virus-Cell Interaction, Institute of Microbiology Paulo de Góes, Federal University of Rio de Janeiro, Av. Carlos Chagas Filho, 373, 21941-902 Rio de Janeiro, Brazil; E-Mail: [email protected] * Author to whom correspondence should be addressed; E-Mail: [email protected]; Tel.: +55-(21)-3936-6743; Fax: +55-(21)-2560-8344. Academic Editor: Kirk Gustafson Received: 1 December 2014 / Accepted: 25 March 2015 / Published: 3 April 2015 Abstract: The marine environment covers almost three quarters of the planet and is where evolution took its first steps. Extremophile microorganisms are found in several extreme marine environments, such as hydrothermal vents, hot springs, salty lakes and deep-sea floors. The ability of these microorganisms to support extremes of temperature, salinity and pressure demonstrates their great potential for biotechnological processes. Hydrolases including amylases, cellulases, peptidases and lipases from hyperthermophiles, psychrophiles, halophiles and piezophiles have been investigated for these reasons. -

Archaeal Distribution and Abundance in Water Masses of the Arctic Ocean, Pacific Sector
Vol. 69: 101–112, 2013 AQUATIC MICROBIAL ECOLOGY Published online April 30 doi: 10.3354/ame01624 Aquat Microb Ecol FREEREE ACCESSCCESS Archaeal distribution and abundance in water masses of the Arctic Ocean, Pacific sector Chie Amano-Sato1, Shohei Akiyama1, Masao Uchida2, Koji Shimada3, Motoo Utsumi1,* 1University of Tsukuba, Tennodai, Tsukuba, Ibaraki 305-8572, Japan 2National Institute for Environmental Studies, Onogawa, Tsukuba, Ibaraki 305-8506, Japan 3Tokyo University of Marine Science and Technology, Konan, Minato-ku, Tokyo 108-8477, Japan ABSTRACT: Marine planktonic Archaea have been recently recognized as an ecologically impor- tant component of marine prokaryotic biomass in the world’s oceans. Their abundance and meta- bolism are closely connected with marine geochemical cycling. We evaluated the distribution of planktonic Archaea in the Pacific sector of the Arctic Ocean using fluorescence in situ hybridiza- tion (FISH) with catalyzed reporter deposition (CARD-FISH) and performed statistical analyses using data for archaeal abundance and geochemical variables. The relative abundance of Thaum - archaeota generally increased with depth, and euryarchaeal abundance was the lowest of all planktonic prokaryotes. Multiple regression analysis showed that the thaumarchaeal relative abundance was negatively correlated with ammonium and dissolved oxygen concentrations and chlorophyll fluorescence. Canonical correspondence analysis showed that archaeal distributions differed with oceanographic water masses; in particular, Thaumarchaeota were abundant from the halocline layer to deep water, where salinity was higher and most nutrients were depleted. However, at several stations on the East Siberian Sea side of the study area and along the North- wind Ridge, Thaumarchaeota and Bacteria were proportionally very abundant at the bottom in association with higher nutrient conditions.