Vol24no7 Pdf-Version.Pdf
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Women, Business and the Law 2020 World Bank Group
WOMEN, BUSINESS AND THE LAW 2020 AND THE LAW BUSINESS WOMEN, WOMEN, BUSINESS AND THE LAW 2020 WORLD BANK GROUP WORLD WOMEN, BUSINESS AND THE LAW 2020 © 2020 International Bank for Reconstruction and Development / The World Bank 1818 H Street NW, Washington, DC 20433 Telephone: 202-473-1000; Internet: www.worldbank.org Some rights reserved 1 2 3 4 23 22 21 20 This work is a product of the staff of The World Bank with external contributions. The findings, interpretations, and conclusions expressed in this work do not necessarily reflect the views of The World Bank, its Board of Executive Directors, or the govern- ments they represent. The World Bank does not guarantee the accuracy of the data included in this work. The boundaries, colors, denominations, and other information shown on any map in this work do not imply any judgment on the part of The World Bank concerning the legal status of any territory or the endorsement or acceptance of such boundaries. Nothing herein shall constitute or be considered to be a limitation upon or waiver of the privileges and immunities of The World Bank, all of which are specifically reserved. Rights and Permissions This work is available under the Creative Commons Attribution 3.0 IGO license (CC BY 3.0 IGO) http://creativecommons.org/ licenses/by/3.0/igo. Under the Creative Commons Attribution license, you are free to copy, distribute, transmit, and adapt this work, including for commercial purposes, under the following conditions: Attribution—Please cite the work as follows: World Bank. 2020. Women, Business and the Law 2020. -

Graduation Ceremonies Geelong October 2017 Published by Deakin University, Geelong VIC 3220 Australia Deakin.Edu.Au
Graduation Ceremonies Geelong October 2017 Published by Deakin University, Geelong VIC 3220 Australia deakin.edu.au © Deakin University 2017 Deakin University CRICOS Provider Code 00113B Printed in Australia by Case Print Management Acknowledgement Trenchers have sharp points and edges that can result in serious injury. For your safety, we do not recommend throwing trenchers. Deakin University’s official photographer will be taking photographs at this graduation event. Your image may be used in Deakin University printed and electronic publications or Deakin social media sites for promotional and educational purposes. This publication is revised annually. The information contained in this edition is accurate as at October 2017. Table of Contents Congratulations 2 Congratulatory message from the Chancellor 2 Congratulatory message from the Vice-Chancellor 3 Welcome to Deakin University Graduations 4 The Graduation Ceremony 6 Acknowledgment of traditional land owners 6 Order of Ceremony 6 The University Mace 7 The Academic Procession 9 Academic Dress 10 Deakin University Ceremonial Dress 10 Deakin University Academic Dress 10 Regalia Colours 11 Deakin Award Appellations 11 Honorary Degree Recipients 12 The Performers 14 The University 16 Alfred Deakin 16 About Deakin University 16 Building on the University’s success 17 Facts about Deakin 18 The Campuses 20 Melbourne Burwood Campus 20 Geelong Waurn Ponds Campus 20 Geelong Waterfront Campus 21 Warrnambool Campus 21 Institute of Koorie Education 22 Get Social! #DeakinGrad 23 Tuesday 3 October 12 pm 25 Tuesday 3 October 6 pm 41 Wednesday 4 October 12 pm 57 Wednesday 4 October 6 pm 77 Thursday 5 October 12 pm 95 Deakin University Alumni Community 118 National Anthem 119 Evacuation Assembly Points 120 Congratulations Congratulatory message from the Chancellor On behalf of the Deakin community, I congratulate you on graduating from Deakin University – your University. -

Ryerson University Spring Graduates
Ryerson University Spring Graduates June 2020 Faculty of Arts 2 Faculty of Communication & Design 11 Faculty of Community Services 21 Faculty of Engineering and Architectural Science 35 Faculty of Science 46 Ted Rogers School of Management 54 Yeates School of Graduate Studies 71 The G. Raymond Chang School of Continuing Education 73 Faculty of Arts Pamela Sugiman Dean Faculty of Arts Janice Fukakusa Chancellor Mohamed Lachemi President and Vice-Chancellor Charmaine Hack Registrar Ryerson Gold Medal Presented to Mayah Obadia Geographic Analysis 2 Faculty of Arts Undergraduate Degree Programs Arts and Contemporary Studies Bachelor of Arts (Honours) *Diana Abo Harmouch Carmen Jajjo *Megumi Noteboom *Sima Rebecca Abrams Leya Jasat Valentina Padure Qeyam Amiri Sophie Johnson *Naiomi Marcia Perera Brodie Barrick Babina Kamalanathan Charlotte Jane Prokopec Rebecca Claire Chen Caroline Susan Kewley Regan Reynolds Erin Tanya Clarke Jessica Laurenza Joshua Ricci *Megan Lisa Devoe Claire Lowenstein Kaitlin Anganie Seepersaud *Manpreet Kaur Dhaliwal *Avigayil Margolis Gabriela Skwarko Tatum Lynn Donovan Sara McArthur Julia Macey Sullivan Faith Raha Giahi *Nadia Celeste McNairn *Helen Gillian Webb Meagan Gove *Mahbod Mehrvarz *Michael Worbanski Salem Habtom Andrew Moon Smyrna Wright *William Hanchar *Liana Gabriella Mortin Calum Jacques Potoula Mozas Criminology Bachelor of Arts (Honours) *Annabelle Adjei *Jenna Anne Giannini Veronica Hiu Lam Lee Stanislav Babinets Albina Glatman Karishma Catherine Lutchman Hela Bakhtari Farah Khaled Gregni Simbiat -

NEWSLETTER 2012 | Vol
Postgraduate Programme PPRE Renewable Energy NEWSLETTER 2012 | Vol. 30 Published by: Carl von Ossietzky University of Oldenburg, Faculty of Physics, Department of Energy and Semiconductor Research, Postgraduate Programme Renewable Energy—PPRE, D - 26111 Oldenburg phone: +49-441-798-3544, fax: +49-441-798-3990, e-mail: [email protected], web: http://www.ppre.de Editorial Team: E. Knagge (in charge), F. Grubitzsch, L. Ibing Typesetting & Layout: Tarek Fakih Printer: Druckzentrum, CvO University Oldenburg - 800 copies Content Editorial 5 News from Oldenburg 25 Years of PPRE 7 30 Years at the Service of Renewable Energies 8 The Future Belongs to Wind Power at University of Oldenburg 11 European Wind Energy Master 12 Continuing Studies Programme Offshore Wind Energy 13 PPRE Online - Premium Online Qualification in Renewable Energy 14 Teaching Award for PPRE Module ‘Rural Energy Supply’ 14 EUREC & PPRE Students Meet Prof. Dan Kammen from World Bank 15 Biogas Compact Workshop 2011 at the University of Oldenburg 16 Excursion to Biogas Plant 18 PPRE Challenge 2011 19 New Coordination of Interdisciplinary Energy Education at University of Oldenburg 20 Cooperations Binational PhD Program Renewable Energy 21 The Developing Sustainability Network (DEVSUS) 22 Research Visit from Partner University in Manaus / Brazil 23 3rd DAAD Network Meeting of the Development-Related Post Graduate Courses 24 Energy and Environmental Management Work at Flensburg University 25 Renewable Energy for Sustainable Development of Indonesia and Germany (RESDIG) 26 2nd -

NEA-Annual-Report-1980.Pdf
National Endowment for the Arts National Endowment for the Arts Washington, D.C. 20506 Dear Mr. President: I have the honor to submit to you the Annual Report of the National Endowment for the Arts and the National Council on the Arts for the Fiscal Year ended September 30, 1980. Respectfully, Livingston L. Biddle, Jr. Chairman The President The White House Washington, D.C. February 1981 Contents Chairman’s Statement 2 The Agency and Its Functions 4 National Council on the Arts 5 Programs 6 Deputy Chairman’s Statement 8 Dance 10 Design Arts 32 Expansion Arts 52 Folk Arts 88 Inter-Arts 104 Literature 118 Media Arts: Film/Radio/Television 140 Museum 168 Music 200 Opera-Musical Theater 238 Program Coordination 252 Theater 256 Visual Arts 276 Policy and Planning 316 Deputy Chairman’s Statement 318 Challenge Grants 320 Endowment Fellows 331 Research 334 Special Constituencies 338 Office for Partnership 344 Artists in Education 346 Partnership Coordination 352 State Programs 358 Financial Summary 365 History of Authorizations and Appropriations 366 Chairman’s Statement The Dream... The Reality "The arts have a central, fundamental impor In the 15 years since 1965, the arts have begun tance to our daily lives." When those phrases to flourish all across our country, as the were presented to the Congress in 1963--the illustrations on the accompanying pages make year I came to Washington to work for Senator clear. In all of this the National Endowment Claiborne Pell and began preparing legislation serves as a vital catalyst, with states and to establish a federal arts program--they were communities, with great numbers of philanthro far more rhetorical than expressive of a national pic sources. -

THE GEOGRAPHY of GALATIA Gal 1:2; Act 18:23; 1 Cor 16:1
CHAPTER 38 THE GEOGRAPHY OF GALATIA Gal 1:2; Act 18:23; 1 Cor 16:1 Mark Wilson KEY POINTS • Galatia is both a region and a province in central Asia Minor. • The main cities of north Galatia were settled by the Gauls in the third cen- tury bc. • The main cities of south Galatia were founded by the Greeks starting in the third century bc. • Galatia became a Roman province in 25 bc, and the Romans established colonies in many of its cities. • Pamphylia was part of Galatia in Paul’s day, so Perga and Attalia were cities in south Galatia. GALATIA AS A REGION and their families who migrated from Galatia is located in a basin in north-cen- Thrace in 278 bc. They had been invited tral Asia Minor that is largely flat and by Nicomedes I of Bithynia to serve as treeless. Within it are the headwaters of mercenaries in his army. The Galatians the Sangarius River (mode rn Sakarya) were notorious for their destructive and the middle course of the Halys River forays, and in 241 bc the Pergamenes led (modern Kızılırmak). The capital of the by Attalus I defeated them at the battle Hittite Empire—Hattusha (modern of the Caicus. The statue of the dying Boğazköy)—was in eastern Galatia near Gaul, one of antiquity’s most noted the later site of Tavium. The name Galatia works of art, commemorates that victo- derives from the twenty thousand Gauls ry. 1 The three Galatian tribes settled in 1 . For the motif of dying Gauls, see Brigitte Kahl, Galatians Re-imagined: Reading with the Eyes of the Vanquished (Minneapolis: Fortress, 2010), 77–127. -

Systematic Position and Taxonomy of Pipistrellus Deserti (Chiroptera: Vespertilionidae)
Mammalia 2015; 79(4): 419–438 Petr Benda*, Tommy Andriollo and Manuel Ruedi Systematic position and taxonomy of Pipistrellus deserti (Chiroptera: Vespertilionidae) Abstract: Pipistrellus deserti is a small, pale-coloured bat DOI 10.1515/mammalia-2014-0024 occurring in the most arid parts of the Sahara, in Morocco, Received February 26, 2014; accepted August 22, 2014; previously published online September 17, 2014 Algeria, Libya, Egypt, and the Sudan, and marginally also in sub-Saharan Africa. Although most authors consider P. deserti as a full species, others regard it as a subspe- cies, or even as a junior synonym of Pipistrellus kuhlii. We Introduction analysed the topotype material of P. deserti from Libya using both morphologic and molecular characters, and The Desert pipistrelle, Pipistrellus deserti, was described compared them with samples from other Saharan coun- by Thomas (1902: 4) as “a small buff-coloured desert ally tries and with P. kuhlii from around the Mediterranean. of P. kuhli” on the basis of a male obtained from Murzuq, The Libyan samples of deserti are morphologically very Fezzan, south-western Libya. Thomas differentiated similar to other populations from arid parts of North his P. deserti from P. kuhlii practically only on the basis Africa (Morocco, Algeria, Egypt, Sudan), but differ mark- of smaller size (e.g., a forearm length 29.5 mm or great- edly in the size of most skull dimensions when compared est skull length 11.6 mm). For a long time, Thomas’ (1902) to P. kuhlii sampled in more mesic areas. However, phy- report remained the only authenticated record of P. -
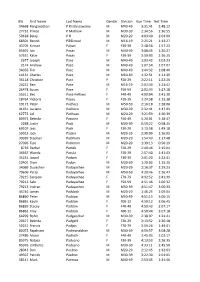
Bib First Name Last Name Gender Division Gun Time Net Time 34668 Ranganathan P Krishnaswamy M M40-49 3:31:41 2:48:12 27732 Princ
Bib First Name Last Name Gender Division Gun Time Net Time 34668 Ranganathan P Krishnaswamy M M40-49 3:31:41 2:48:12 27732 Prince P Mathew M M30-39 2:34:56 1:36:55 50628 Balaji P R M M20-29 4:03:40 2:04:09 68501 Benoit Pécoud M M16-19 2:25:21 1:13:27 43705 Kinnari Pabari F F30-39 2:48:56 1:57:23 85691 Jan Pacas M M40-49 3:08:45 1:20:27 57631 Katie Pacas F F30-39 3:59:03 2:36:25 2677 Joseph Pace M M40-49 1:03:42 1:03:23 2174 Andrew Pace M M40-49 1:07:54 1:07:07 34056 Tim Pace M M40-49 1:44:32 1:08:40 14131 Charles Pace M M60-69 1:32:51 1:21:39 36118 Christine Pace F F20-29 2:22:11 1:22:26 24221 Ben Pace M M16-19 2:02:34 1:23:42 26478 Susan Pace F F50-59 2:02:33 1:27:18 55511 Bec Pace-Fellows F F40-49 4:00:04 2:41:30 18764 Victoria Pacey F F20-29 1:24:58 1:13:38 19172 Peter Pachacz M M50-59 2:16:18 1:28:06 41451 Luciano Pacheco M M30-39 2:32:41 1:47:03 62772 Lal Pachuau M M20-29 3:21:59 1:26:39 80972 Belinda Pacione F F40-49 5:20:01 3:18:47 2268 Justin Pack M M30-39 0:59:22 0:58:25 69107 Jess Pack F F20-29 3:13:58 1:49:18 59013 Jack Packer M M20-29 2:39:09 1:16:05 39939 Stephen Packham M M20-29 1:54:43 1:17:48 27096 Tipo Packman M M20-29 1:39:13 0:56:29 8736 Rachel Pacleb F F20-29 2:40:46 2:15:04 34467 Wanda Pacula F F20-29 2:57:40 1:41:43 35224 Jaspal Padam F F30-39 2:01:20 1:23:41 10401 Dion Padan M M30-39 1:29:03 1:15:25 34069 Duvashen Padayachee M M20-29 2:16:37 1:29:23 79606 Perry Padayachee M M50-59 4:20:46 2:26:47 79215 Sarojeni Padayachee F F70-79 4:32:52 2:41:39 79212 Salo Padayachee F F50-59 4:51:46 3:00:32 79213 Indiran Padayachee -

Bark at Park’ Festival by Everett Rosenfeld ‘09 May Day, the Lower School’S Liss, Selected the Music for the Event
POSTSCRIPT The Park School Brooklandville, MD May 28, 2004 Volume LXIV Issue No. 9 Parents’ Healthy Food Committee bans soda sales on campus by Sarah Dunn ‘06 Starting next year, Park will ban daily nutritional requirements. It is stat- to more healthy drinks. At Oldfields, healthier foods in schools is happening the sale of sodas on campus. This move ed directly in the proposal that, “health many other junk foods have been elimi- all over the country. In New York City is the result of an April 14 proposal made related behaviors are established early in nated as well. Almost 95% of what is sold and Los Angeles public schools, sodas by the Park Healthy Foods Committee, life” and the Committee believes that in its vending machines are healthy were banned from vending machines. In a group that consists of faculty and par- Philadelphia public schools, all soda sales ents. The proposal stated that the school were eliminated. Other states taking a should not profit from the sale of such stand on the foods sold in their cafeteri- unhealthy drinks and that they should not as include Texas, South Carolina, New be sold on school grounds. The Park Hampshire, Washington, California, and Parents’ Association approved the plan; Minnesota. Dr. David Jackson, Head of School, and Next year, Park will discontin- Caleb Karpay ‘04, former Upper School ue its vending machine soda sales as President, also supported it. well. Unlike St. Paul’s, students will be Deirdre Smith, a parent and a permitted to bring sodas on campus, but member of the committee, explained that the school does not wish to profit from the number one disease among teenag- them. -

Inscriptions from Northwest Pisidia 3
Habelt-Verlag · Bonn Epigraphica Anatolica 48 (2015) 1–85 IINSCRIPTIONSNSCRIPTIONS FFROMROM NNORTHWESTORTHWEST PISIDIAPISIDIA The inscriptions published below were all found or studied as a part of the Isparta Archaeologi- cal Survey from 2009 to 2015,1 thirty-one of which are published here for the first time.2 1 I am particularly grateful to Director of the Isparta Archaeological Survey (IAS), Bilge Hürmüzlü, for all her support and encouragement. Thanks also go to Andrea De Giorgi (co-Director of the IAS until 2011), as well as to the T. C. Kültür ve Turizm Bakanlığı for the survey and museum permits and financial support, to the min- istry representatives in 2009–2015, and to Süleyman Demirel Üniversitesi for providing support, including the IAS’s Survey House. Also special thanks go to İlhan Güceren and Mustafa Akaslan of the Isparta Museum and Hacı Ali Ekinci of the Burdur Museum for granting access to the collections, and to the Case Western Reserve University’s College of Arts and Sciences, Department of Classics, and the Baker Nord Center for the Humanities for their financial support. 2 On the research of the Isparta Archaeological Survey, see bibliography cited by Iversen 2012, p. 103, n. 2. Since then, also see B. Hürmüzlü and P. Iversen, Notes on Cultural Interaction in Northwest Pisidia in the Iron Age, in N. Chr. Stampolidis, Ç. Maner, K. Kopanias (eds), NOSTOI: Indigenous Culture, Migration, and Integration in the Aegean Islands and Western Anatolia During the Late Bronze and Early Iron Ages (Istanbul 2015), pp. 531–537; A. De Giorgi, Between Continuity and Change: Northern Pisidia Through Classical and Late Antiquity, MDAI(I) 64 (2014), pp. -

NEWSLETTER No
pre_blau.jpg Postgraduate Programme neu_carl.pdf RENEWABLE ENERGY NEWSLETTER No. 1/2002 – Vol 21 EDITORIAL Dear Reader—spring has finally arrived and sunlight is filtering through the first leaves of some bushes at the Wechloy Campus here in Olden- burg. This newsletter was intended to be published in fall 2001. Edu Knagge’s many duties (see last newsletter!) prevented him from work- ing on this issue, so I took over and started work on the collection of e-mails and articles collected by our friend and colleague only after the Winter Term 2002 was finished. This explains the delay and I learned (by doing) what time and effort it requires to complete an issue of the PPRE Newsletter and only after some weeks of sifting thorough quite a few e-mails and some MB of attached files things got into place. So, what do you have to expect from this issue? Well, we will try and present some News from Oldenburg, keep you informed about messages that arrived from the PPRE Alumni and then you might like some articles sent from some of your former colleagues. An overview on publications and useful web sites will complete the newsletter – at the end like always an updated list of alumni and their e-mail addresses. In 2002 PPRE will pass 15 years of programme and the number of ab- solvents will pass the 0 mark. The curriculum will be changed due to changes in the field of renewable energy. Happy reading and good wishes from Oldenburg signa_kb.jpg CONTENTS EDITORIAL . 1 NEWS FROM OLDENBURG . -

Displaying the Res Gestae of Augustus: a Monument of Imperial
Displaying the Res Gestae of Augustus: A Monument of Imperial Image for All Author(s): Suna Güven Reviewed work(s): Source: Journal of the Society of Architectural Historians, Vol. 57, No. 1 (Mar., 1998), pp. 30- 45 Published by: University of California Press on behalf of the Society of Architectural Historians Stable URL: http://www.jstor.org/stable/991403 . Accessed: 26/01/2012 09:51 Your use of the JSTOR archive indicates your acceptance of the Terms & Conditions of Use, available at . http://www.jstor.org/page/info/about/policies/terms.jsp JSTOR is a not-for-profit service that helps scholars, researchers, and students discover, use, and build upon a wide range of content in a trusted digital archive. We use information technology and tools to increase productivity and facilitate new forms of scholarship. For more information about JSTOR, please contact [email protected]. University of California Press and Society of Architectural Historians are collaborating with JSTOR to digitize, preserve and extend access to Journal of the Society of Architectural Historians. http://www.jstor.org Displayingthe Res Gestae of Augustus A Monumentof ImperialImage for All most literal sense, it is basically a catalogue of the achieve- SUNA GLIVEN,Middle East TechnicalUniversity, Ankara ments of the Divine Augustus. Looking at it another way, we R oman inscriptions, and others, are usually studied as could say that it starts off as an altruistic record of the first extual documents that recordhistory. In this traditional Roman emperor and his performance designed by a "memory approach, specialists in epigraphy literally translate the written entrepreneur," to use a term coined by James Young.4 Follow- text so that it becomes, on its own, the veritable evidence for ing the last injunction of the emperor, who died on 19 August what it records.