Eliminating Blood Oncogenic Exosomes Into the Small Intestine with Aptamer-Functionalized Nanoparticles
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

World Higher Education Database Whed Iau Unesco
WORLD HIGHER EDUCATION DATABASE WHED IAU UNESCO Página 1 de 438 WORLD HIGHER EDUCATION DATABASE WHED IAU UNESCO Education Worldwide // Published by UNESCO "UNION NACIONAL DE EDUCACION SUPERIOR CONTINUA ORGANIZADA" "NATIONAL UNION OF CONTINUOUS ORGANIZED HIGHER EDUCATION" IAU International Alliance of Universities // International Handbook of Universities © UNESCO UNION NACIONAL DE EDUCACION SUPERIOR CONTINUA ORGANIZADA 2017 www.unesco.vg No paragraph of this publication may be reproduced, copied or transmitted without written permission. While every care has been taken in compiling the information contained in this publication, neither the publishers nor the editor can accept any responsibility for any errors or omissions therein. Edited by the UNESCO Information Centre on Higher Education, International Alliance of Universities Division [email protected] Director: Prof. Daniel Odin (Ph.D.) Manager, Reference Publications: Jeremié Anotoine 90 Main Street, P.O. Box 3099 Road Town, Tortola // British Virgin Islands Published 2017 by UNESCO CENTRE and Companies and representatives throughout the world. Contains the names of all Universities and University level institutions, as provided to IAU (International Alliance of Universities Division [email protected] ) by National authorities and competent bodies from 196 countries around the world. The list contains over 18.000 University level institutions from 196 countries and territories. Página 2 de 438 WORLD HIGHER EDUCATION DATABASE WHED IAU UNESCO World Higher Education Database Division [email protected] -

An Chengri an Chengri, Male, Born in November, 1964.Professor. Director
An Chengri , male, born in November, 1964.Professor. Director of Institute of International Studies, Department of Political Science, School of philosophy and Public Administration,Heilongjiang University. Ph. D student of Japanese politics and Diplomacy History, NanKai University,2001.Doctor(International Relations History), Kokugakuin University,2002. Research Orientation: Japanese Foreign Relations, International Relation History in East Asia Publications: Research on contemporary Japan-South Korea Relations(China Social Science Press,October,2008);International Relations History of East Asia(Jilin Science Literature Press,March,2005) Association: Executive Director of China Institute of Japanese History , Director of China Society of Sino-Japanese Relations History Address: No.74 Xuefu Road, Nangang District, Haerbin, Heilongjiang, Department of Political Science, School of philosophy and Public Administration,Heilongjiang University. Postcode: 150080 An shanhua , Female, born in July,1964. Associate Professor, School of History, Dalian University. Doctor( World History),Jilin University,2007. Research Orientation: Modern and contemporary Japanese History, Japanese Foreign Relations, Political Science Publications: Comparative Studies on World Order View of China Korea and Japan and their Diplomatic in Modern Time ( Japanese Studies Forum , Northeast Normal University, 2006); Analysis of Japan's anti-system ideology towards the international system ( Journal of Changchun University of Science and Technology , Changchun University,2006) -

A Complete Collection of Chinese Institutes and Universities For
Study in China——All China Universities All China Universities 2019.12 Please download WeChat app and follow our official account (scan QR code below or add WeChat ID: A15810086985), to start your application journey. Study in China——All China Universities Anhui 安徽 【www.studyinanhui.com】 1. Anhui University 安徽大学 http://ahu.admissions.cn 2. University of Science and Technology of China 中国科学技术大学 http://ustc.admissions.cn 3. Hefei University of Technology 合肥工业大学 http://hfut.admissions.cn 4. Anhui University of Technology 安徽工业大学 http://ahut.admissions.cn 5. Anhui University of Science and Technology 安徽理工大学 http://aust.admissions.cn 6. Anhui Engineering University 安徽工程大学 http://ahpu.admissions.cn 7. Anhui Agricultural University 安徽农业大学 http://ahau.admissions.cn 8. Anhui Medical University 安徽医科大学 http://ahmu.admissions.cn 9. Bengbu Medical College 蚌埠医学院 http://bbmc.admissions.cn 10. Wannan Medical College 皖南医学院 http://wnmc.admissions.cn 11. Anhui University of Chinese Medicine 安徽中医药大学 http://ahtcm.admissions.cn 12. Anhui Normal University 安徽师范大学 http://ahnu.admissions.cn 13. Fuyang Normal University 阜阳师范大学 http://fynu.admissions.cn 14. Anqing Teachers College 安庆师范大学 http://aqtc.admissions.cn 15. Huaibei Normal University 淮北师范大学 http://chnu.admissions.cn Please download WeChat app and follow our official account (scan QR code below or add WeChat ID: A15810086985), to start your application journey. Study in China——All China Universities 16. Huangshan University 黄山学院 http://hsu.admissions.cn 17. Western Anhui University 皖西学院 http://wxc.admissions.cn 18. Chuzhou University 滁州学院 http://chzu.admissions.cn 19. Anhui University of Finance & Economics 安徽财经大学 http://aufe.admissions.cn 20. Suzhou University 宿州学院 http://ahszu.admissions.cn 21. -

Exploring a New Chinese Model of Tourism and Hospitality Education
2017 International Conference on Energy, Environment and Sustainable Development (EESD 2017) ISBN: 978-1-60595-452-3 Exploring a New Chinese Model of Tourism and Hospitality Education: Lessons Learned from American Counterparts Jin-lin WU1, Zong-qing ZHOU2, Hong-bin XIE3 and Fei CHENG3 1Department of Tourism, Minjiang University, China 2College of Hospitality Management, Niagara University, USA 3Institute of Tourism Research, Fujian Normal University, China Keywords: College-enterprise cooperation, Tourism talents training mode, Experiences of America, China. Abstract. College-enterprise cooperation has become one of the important modes of tourism management education in colleges and universities of China. This article probed on experiences of college-enterprise cooperation in tourism colleges of America, and then presented the situation of college-enterprise cooperation in tourism college of China, and further analyzed its main reasons. Referring to the experiences of America, this article discussed personnel training modes which conform to both the international popular training practices and Chinese characteristics in order to train tourism talents. Introduction College-enterprise cooperation is a kind of "win-win" cooperative mode for colleges and enterprises to train talents, which means the students acquire their theoretical knowledge in college and practical experiences in enterprise, meanwhile the colleges and enterprises share common human resources and information. At present, the cooperation between colleges and enterprises has become one of the important modes for training professional talents of tourism management in colleges of China. Compared to America, which have rich experiences on college-enterprise cooperation, the tourism education in China started later with problems about the depth of college-enterprise cooperation and the modes of training students' comprehensive ability. -

University of Leeds Chinese Accepted Institution List 2021
University of Leeds Chinese accepted Institution List 2021 This list applies to courses in: All Engineering and Computing courses School of Mathematics School of Education School of Politics and International Studies School of Sociology and Social Policy GPA Requirements 2:1 = 75-85% 2:2 = 70-80% Please visit https://courses.leeds.ac.uk to find out which courses require a 2:1 and a 2:2. Please note: This document is to be used as a guide only. Final decisions will be made by the University of Leeds admissions teams. -

Conquering Gender Stereotype Threat in “Digit Sports”: Effects of Gender Swapping on Female Players' Continuous Participation Intention in Esports
Hindawi Scientific Programming Volume 2020, Article ID 8818588, 7 pages https://doi.org/10.1155/2020/8818588 Research Article Conquering Gender Stereotype Threat in “Digit Sports”: Effects of Gender Swapping on Female Players’ Continuous Participation Intention in ESports Lujie Hao,1,2 Qinghua Lv,1 Xiaosan Zhang ,3 Qingquan Jiang ,4 Songxian Liu,4 and Lin Ping5 1School of Business, Huaqiao University, Quanzhou 362021, China 2Economics and Business Administration Institute, Fuzhou University of Foreign Studies and Trade, Fuzhou 350202, China 3Research Centre for Belt & Road Financial and Economic Development, Xiamen National Accounting Institute, Xiamen 361005, China 4School of Economics & Management, Xiamen University of Technology, Xiamen 361024, China 5Chinese Entrepreneurship Research Center, Minjiang University, Fuzhou 350108, China Correspondence should be addressed to Xiaosan Zhang; [email protected] and Qingquan Jiang; [email protected] Received 14 April 2020; Revised 30 May 2020; Accepted 16 June 2020; Published 6 July 2020 Academic Editor: Chenxi Huang Copyright © 2020 Lujie Hao et al. .is is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. As a sportification form of human-computer interaction, eSports is facing great gender stereotype threat and causing female players’ withdraw. .is study aims to investigate the relationship between gender-swapping and females’ continuous -

Millet Arrived in the South China Coast Around 5,500 Years Ago
EGU21-8311, updated on 27 Sep 2021 https://doi.org/10.5194/egusphere-egu21-8311 EGU General Assembly 2021 © Author(s) 2021. This work is distributed under the Creative Commons Attribution 4.0 License. Millet arrived in the South China Coast around 5,500 years ago Jinqi Dai1,2,3, Xipeng Cai4, Jianhui Jin1,2,3, Wei Ge5, Yunming Huang6, Wei Wu7,8, Taoqin Xia4, Fusheng Li4, and Xinxin Zuo1,2,3 1Key Laboratory for Humid Subtropical Eco-geographical Processes of the Ministry of Education, Fujian Normal University, Fuzhou, China 2School of Geographical Sciences, Fujian Normal University, Fuzhou, China 3Center for Environmental Archaeology in Southeast China, Fujian Normal University, Pingtan, China 4Fuzhou Municipal Institute of Archaeology, Fuzhou, China 5School of Humanities, Xiamen University, Xiamen, China 6School of Humanities, Minjiang University, Fuzhou, China 7College of Architecture and Urban Planning, Fujian University of Technology, Fuzhou, China 8Tanshishan Site Museum, Fuzhou, China Crop dispersal has long been recognised as an important topic in agricultural archaeology and food globalisation. One of most pressing questions facing archaeologists is determining when and where millet arrived in the South China Coast. Our study focused on the millet phytoliths remains from three Neolithic sites in southeast coastal Fujian. Multiple dating methods, including charred carbon dating, phytolith carbon dating, and optically stimulated luminescence were used to construct the chronologies of the sites. The dating results showed that BTS was initially occupied at approximately 5,500 cal a BP. The millet phytoliths recovered in this study are likely the earliest millet remains found in Fujian, suggesting that millet arrived in the South China Coast at least 5,500 years ago. -

Label-Free Multiphoton Imaging to Assess Neoadjuvant Therapy
Int. J. Biol. Sci. 2020, Vol. 16 1376 Ivyspring International Publisher International Journal of Biological Sciences 2020; 16(8): 1376-1387. doi: 10.7150/ijbs.41579 Research Paper Label-free multiphoton imaging to assess neoadjuvant therapy responses in breast carcinoma Lianhuang Li1*, Zhonghua Han2*, Lida Qiu1 3*, Deyong Kang4, Zhenlin Zhan1, Haohua Tu5, and Jianxin Chen1 1. Key Laboratory of OptoElectronic Science and Technology for Medicine of Ministry of Education, Fujian Provincial Key Laboratory for Photonics Technology, Fujian Normal University, Fuzhou 350007, P. R. China 2. Department of Breast Surgery, Fujian Medical University Union Hospital, Fuzhou 350001, P. R. China 3. College of Physics and Electronic Information Engineering, Minjiang University, Fuzhou 350108, P. R. China 4. Department of Pathology, Fujian Medical University Union Hospital, Fuzhou 350001, P. R. China 5. Beckman Institute for Advanced Science and Technology, University of Illinois at Urbana-Champaign, Urbana, IL 61801, USA * These authors contributed equally to this work. Corresponding authors: Lianhuang Li and Jianxin Chen, College of Photonic and Electronic Engineering, Fujian Normal University, Fuzhou 350007, P. R. China; E-mail: [email protected] and [email protected]; Or Zhonghua Han, Department of Breast Surgery, Fujian Medical University Union Hospital, Fuzhou 350001, P. R. China; E-mail: [email protected]. © The author(s). This is an open access article distributed under the terms of the Creative Commons Attribution License (https://creativecommons.org/licenses/by/4.0/). See http://ivyspring.com/terms for full terms and conditions. Received: 2019.10.31; Accepted: 2020.02.02; Published: 2020.02.21 Abstract Neoadjuvant chemotherapy has been used increasingly in patients with early-stage or locally advanced breast carcinoma, and has been recommended as a general approach in locally advanced-stage diseases. -

Large-Scale Tumor-Associated Collagen Signatures Identify High-Risk Breast Cancer Patients
Large-scale tumor-associated collagen signatures identify high-risk breast cancer patients Gangqin Xi1†, Wenhui Guo2†, Deyong Kang3†, Jianli Ma4†, Fangmeng Fu2, Lida Qiu1,5, Liqin Zheng1, Jiajia He1, Na Fang1,6, Jianhua Chen1,7, Jingtong Li8, Shuangmu Zhuo1, Xiaoxia Liao9, Haohua Tu10, Lianhuang Li1, Qingyuan Zhang8, Chuan Wang2, Stephen A. Boppart10, and Jianxin Chen1 1. Key Laboratory of OptoElectronic Science and Technology for Medicine of Ministry of Education, Fujian Provincial Key Laboratory of Photonics Technology, Fujian Normal University, Fuzhou, China 2. Breast Surgery Ward, Department of General Surgery, Fujian Medical University Union Hospital, Fuzhou, China 3. Department of Pathology, Fujian Medical University Union Hospital, Fuzhou, China 4. Department of Radiation Oncology, Harbin Medical University Cancer Hospital, Harbin, China 5. College of Physics and Electronic Information Engineering, Minjiang University, Fuzhou, China 6. Department of Ophthalmology and Optometry, Fujian Medical University, Fuzhou, China 7. College of Life Science, Fujian Normal University, Fuzhou, China 8. Department of Medical Oncology, Harbin Medical University Cancer Hospital, Harbin, China 9. National Center for Supercomputing Applications, University of Illinois at Urbana-Champaign, Urbana, USA 10. Beckman Institute for Advanced Science and Technology, University of Illinois at Urbana-Champaign, Urbana, USA †These authors contributed equally to this work. Corresponding authors: Lianhuang Li, College of Photonic and Electronic Engineering, Fujian Normal University, Fuzhou 350007, China, Telephone number: 86-591-83465818, E-mail: [email protected]; Qingyuan Zhang, Department of Medical Oncology, Harbin Medical University Cancer Hospital, Harbin 150081, China, Telephone number: 86-13313612989, E-mail: [email protected]; Chuan Wang, Breast Surgery Ward, Department of General Surgery, Fujian Medical University Union Hospital, Fuzhou 350001, China, Telephone number: 86-13515020716, E-mail: [email protected]; Stephen A. -
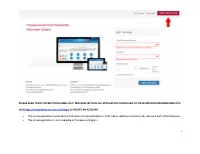
1 Please Read These Instructions Carefully
PLEASE READ THESE INSTRUCTIONS CAREFULLY. MISTAKES IN YOUR CSC APPLICATION COULD LEAD TO YOUR APPLICATION BEING REJECTED. Visit http://studyinchina.csc.edu.cn/#/login to CREATE AN ACCOUNT. • The online application works best with Firefox or Internet Explorer (11.0). Menu selection functions may not work with other browsers. • The online application is only available in Chinese and English. 1 • Please read this page carefully before clicking on the “Application online” tab to start your application. 2 • The Program Category is Type B. • The Agency No. matches the university you will be attending. See Appendix A for a list of the Chinese university agency numbers. • Use the + by each section to expand on that section of the form. 3 • Fill out your personal information accurately. o Make sure to have a valid passport at the time of your application. o Use the name and date of birth that are on your passport. Use the name on your passport for all correspondences with the CLIC office or Chinese institutions. o List Canadian as your Nationality, even if you have dual citizenship. Only Canadian citizens are eligible for CLIC support. o Enter the mailing address for where you want your admission documents to be sent under Permanent Address. Leave Current Address blank. Contact your home or host university coordinator to find out when you will receive your admission documents. Contact information for you home university CLIC liaison can be found here: http://clicstudyinchina.com/contact-us/ 4 • Fill out your Education and Employment History accurately. o For Highest Education enter your current degree studies. -

Conference Program
2017 4th International Conference on Information Science and Control Engineering ICISCE 2017 Conference Program Dolton Resort Tongshenhu Hotel - Changsha July 21-23, 2017, Changsha, China Technically Co-Sponsored by IEEE Computer Society, USA Sponsored by Hunan University of Humanities, Science and Technology, China University of Technology Sydney, Australia Hunan University, China Wayne State University, USA Minjiang University, China Beijing University of Civil Engineering and Architecture, China Xiamen University, China China Jiliang University, China Iwate Prefectural University, Japan Message from the ICISCE 2017 General Chairs ICISCE 2017 is the 4th International Conference on Information Science and Control Engineering, will be held from July 21-23, 2017, in Changsha, China. The aim of the ICISCE 2017 is to provide an international conference for scientific research on Information Science and Control Engineering. ICISCE 2017 is Technically Co-Sponsored by IEEE Computer Society, and sponsored by Hunan University of Humanities, Science and Technology, and Co-sponsored by University of Technology Sydney, Hunan University, Wayne State University, Minjiang University, Beijing University of Civil Engineering and Architecture, Xiamen University, China Jiliang University, Iwate Prefectural University. ICISCE 2017 is the next event in a series of highly successful the International conference on Information Science and Control Engineering, ICISCE 2012 (December 7-9, 2012, Shenzhen, China), ICISCE 2015 (April 24-26, 2015, Shanghai, China), ICISCE 2016 (July 8-10, 2016, Beijing, China). We would like to express our sincere thanks to the Program Chairs: Shaozi Li (Xiamen University, China), Ying Dai (Iwate Prefectural University, Japan), Daniel Neagu (University of Bradford, UK), all program committee members and all the additional reviewers for their valuable efforts in the review process, which helped us to guarantee the highest quality of the selected papers for the conference. -

99Mtc-MIBI Positive of Cervical Ectopic Thymoma Case Report and Literature Review
2019 10th International Conference on Information Technology in Medicine and Education (ITME 2019) Qingdao, China 23 – 25 August 2019 IEEE Catalog Number: CFP1953E-POD ISBN: 978-1-7281-3919-7 Copyright © 2019 by the Institute of Electrical and Electronics Engineers, Inc. All Rights Reserved Copyright and Reprint Permissions: Abstracting is permitted with credit to the source. Libraries are permitted to photocopy beyond the limit of U.S. copyright law for private use of patrons those articles in this volume that carry a code at the bottom of the first page, provided the per-copy fee indicated in the code is paid through Copyright Clearance Center, 222 Rosewood Drive, Danvers, MA 01923. For other copying, reprint or republication permission, write to IEEE Copyrights Manager, IEEE Service Center, 445 Hoes Lane, Piscataway, NJ 08854. All rights reserved. *** This is a print representation of what appears in the IEEE Digital Library. Some format issues inherent in the e-media version may also appear in this print version. IEEE Catalog Number: CFP1953E-POD ISBN (Print-On-Demand): 978-1-7281-3919-7 ISBN (Online): 978-1-7281-3918-0 ISSN: 2474-381X Additional Copies of This Publication Are Available From: Curran Associates, Inc 57 Morehouse Lane Red Hook, NY 12571 USA Phone: (845) 758-0400 Fax: (845) 758-2633 E-mail: [email protected] Web: www.proceedings.com 2019 10th International Conference on Information Technology in Medicine and Education (ITME) ITME 2019 Table of Contents Message from the ITME 2019 General Chairs xxiii Message from