Cholecseg8k: a Semantic Segmentation Dataset for Laparoscopic Cholecystectomy Based on Cholec80
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

SPLANCHNOLOGY Part I. Digestive System (Пищеварительная Система)
КАЗАНСКИЙ ФЕДЕРАЛЬНЫЙ УНИВЕРСИТЕТ ИНСТИТУТ ФУНДАМЕНТАЛЬНОЙ МЕДИЦИНЫ И БИОЛОГИИ Кафедра морфологии и общей патологии А.А. Гумерова, С.Р. Абдулхаков, А.П. Киясов, Д.И. Андреева SPLANCHNOLOGY Part I. Digestive system (Пищеварительная система) Учебно-методическое пособие на английском языке Казань – 2015 УДК 611.71 ББК 28.706 Принято на заседании кафедры морфологии и общей патологии Протокол № 9 от 18 апреля 2015 года Рецензенты: кандидат медицинских наук, доцент каф. топографической анатомии и оперативной хирургии КГМУ С.А. Обыдённов; кандидат медицинских наук, доцент каф. топографической анатомии и оперативной хирургии КГМУ Ф.Г. Биккинеев Гумерова А.А., Абдулхаков С.Р., Киясов А.П., Андреева Д.И. SPLANCHNOLOGY. Part I. Digestive system / А.А. Гумерова, С.Р. Абдулхаков, А.П. Киясов, Д.И. Андреева. – Казань: Казан. ун-т, 2015. – 53 с. Учебно-методическое пособие адресовано студентам первого курса медицинских специальностей, проходящим обучение на английском языке, для самостоятельного изучения нормальной анатомии человека. Пособие посвящено Спланхнологии (науке о внутренних органах). В данной первой части пособия рассматривается анатомическое строение и функции системы в целом и отдельных органов, таких как полость рта, пищевод, желудок, тонкий и толстый кишечник, железы пищеварительной системы, а также расположение органов в брюшной полости и их взаимоотношения с брюшиной. Учебно-методическое пособие содержит в себе необходимые термины и объём информации, достаточный для сдачи модуля по данному разделу. © Гумерова А.А., Абдулхаков С.Р., Киясов А.П., Андреева Д.И., 2015 © Казанский университет, 2015 2 THE ALIMENTARY SYSTEM (systema alimentarium/digestorium) The alimentary system is a complex of organs with the function of mechanical and chemical treatment of food, absorption of the treated nutrients, and excretion of undigested remnants. -

Gallbladder Diseases ♧ 1- Cholelithiasis & Cholecystitis = Inflammation of GB
10 Alia Abbadi Ibrahim Elhaj Ibrahim Elhaj Mohammad Al-Mohtaseb Anything This lecture will be about the liver and the gallbladder. Enjoy underlined in this sheet is in the ♧ The surfaces of the liver ♧ slides and was not mentioned by 1) Postero - inferior surface= visceral surface (explained in the previous lecture) the professor. 2) Anterior surface 3) Posterior surface 4) Superior surface = Diaphragmatic surface 5) Right surface 《1》Visceral surface let's go over the relations of this surface quickly ▪ Inferior vena cava (I.V.C) ▪ The gallbladder ▪ Esophagus → with esophageal groove of liver ▪ Stomach → with gastric impression ▪ Duodenum → Duodenal impression (near the neck of the gallbladder) ▪ Right colic flexure → Colic impression ▪ Right kidney → Renal impression ▪ Right suprarenal gland → Suprarenal impression ▪ Porta hepatis (bile duct, hepatic artery, portal vein, lymph nodes, ANS fibers) ▪ Ligamentum venosum & lesser omentum → there is a fissure for both ▪ Ligamentum teres ▪ Tubular omentum (tuber omentale related to pancreas) 《2》Anterior surface Relations of the liver anteriorly: ▪ Diaphragm (it extends to cover part of the anterior surface of the liver) ▪ Right & left pleura and lungs (the diaphragm separates between them and the liver) ▪ Costal cartilage ▪ Xiphoid process ▪ Anterior abdominal wall 《3》Posterior surface ▪ Diaphragm ▪ Right kidney and the supra-renal gland ▪ Transverse colon (right colic flexure) ▪ Duodenum ▪ Gall bladder ▪ I.V.C ▪ Esophagus ▪ Fundus(base) of stomach 《4》Superior surface ▪ Diaphragm ▪ Cut edge of the falciform ligament ▪ Ligamentum teres ▪ The left and right triangular ligaments ▪ Fundus of gall bladder ▪ The cut edges of the superior and inferior parts of the coronary ligament ▪ I.V.C and the 3 hepatic veins (right, left & middle) → passing through a groove ⇒ The 3 hepatic veins drain into I.V.C ⇒ The caudate lobe of the liver more or less is wrapping around the groove of the IVC ⇒ Right & left liver lobes and the bare area of the liver can be seen from the superior view. -

Video Article Comparative Surgical Resection of Ligamentum Teres
Video Article Comparative surgical resection of ligamentum teres hepatis both in cadaveric model and ovarian cancer patient Selçuk et al. Comparative resection of ligamentum teres hepatis İlker Selçuk1, Zehra Öztürk Başarır1, Nurian Ohri2, Bertan Akar3, Eray Çalışkan4, Tayfun Güngör1 1Department of Gynecologic Oncology, Health Sciences University, Zekai Tahir Burak Woman’s Health Training and Research Hospital, Ankara, Turkey 2Department of General Surgery, Division of Surgical Oncology, Ankara University Faculty of Medicine, Ankara, Turkey 3Department of Obstetrics and Gynecology, Bahçeşehir University Faculty of Medicine, İstanbul, Turkey 4Department of Obstetrics and Gynecology, İstinye University, WM Medical Park Hospital, Kocaeli, Turkey Adress for Correspondence: İlker Selçuk Phone: +90 312 306 50 00 e.mail: [email protected] ORCID ID: orcid.org/0000-0003-0499-5722 DOI: 10.4274/jtgga.2018.0135 Received: 1 October, 2018 Accepted: 1 November, 2018 Abstract Resection of all tumor implants with the aim of maximal cytoreduction is the main predictor of overall survival in ovarian carcinoma. However, there are high risk sites of tumor recurrence, andProof perihepatic region especially the point where ligamentum teres hepatis enters the liver parenchyma under the hepatic bridge (pont hepatique) is one of them. This video demonstrates the resection of ligamentum teres hepatis both in a cadaveric model and ovarian cancer patient. (J Turk Ger Gynecol Assoc 2019; 20: xxxxx) Keywords: Pont hepatique, umbilical ligament, liver, ovarian cancer, cytoreduction Introduction The falciforme ligament divides the liver into right and left lobes on the antero-superior part of portoumbilical fissure in which ligamentum teres hepatis (umbilical ligament of liver/round ligament of liver) attaches to the visceral surface. -

C:\Users\Admin.Dora1-PC\Desktop\Dr Pawan XPS\26-28 IJVA.Xps
Indian Journal of Veterinary Anatomy 32 (1): 26-28, June 2020 Morphology of the Liver of Asian Elephant (Eelephas maximus indicus) C.V. Rajani1*, H.S. Patki2, K.P. Surjith3, S. Patgiri4, P.M. Deepa5 and D. Abraham6 Department of Veterinary Anatomy and Histology College of Veterinary and Animal Sciences, Pookode Wayanad-673 576 (Kerala) Received: 05 June 2020; Accepted: 06 June 2020 ABSTRACT The study was conducted on liver collected from an elephant calf and 5 adult domesticated Asian elephants. After recording gross morphological features, liver samples were processed for routine histological procedures. Liver had four borders viz., dorsal, ventral, left lateral and right lateral. Caudal vena cava crossed the dorsal border and the ventral border had a deep umbilical fissure. Parietal surface presented straight line of attachment of Falciform ligament and an inverted arc-like coronary ligament. Visceral surface had a wide hepatic porta and attachment of lesser omentum. Liver had two chief lobes: a larger right lobe and an undivided left lobe. The round ligament was attached to the right medial lobe. Liver did not adhere to right kidney. Gall bladder was absent. Liver showed dense Glisson’s capsule. The hepatic lobules were indistinctly delineated and irregularly hexagonal. Radiating, branching laminae of hepatocytes and intervening anastomosing sinusoids constituted the parenchyma. Small squamous endothelial cells and large Kupffer cells lined the sinusoids. Portal triads were observed at the periphery of the lobule. The present study established the morphological and histological characteristics of liver in Asian elephant. Key words: Asian elephant, Morphology, Histology, Liver, Hepatocytes Anatomical features on viscera of wild animals are diaphragm. -

26 April 2010 TE Prepublication Page 1 Nomina Generalia General Terms
26 April 2010 TE PrePublication Page 1 Nomina generalia General terms E1.0.0.0.0.0.1 Modus reproductionis Reproductive mode E1.0.0.0.0.0.2 Reproductio sexualis Sexual reproduction E1.0.0.0.0.0.3 Viviparitas Viviparity E1.0.0.0.0.0.4 Heterogamia Heterogamy E1.0.0.0.0.0.5 Endogamia Endogamy E1.0.0.0.0.0.6 Sequentia reproductionis Reproductive sequence E1.0.0.0.0.0.7 Ovulatio Ovulation E1.0.0.0.0.0.8 Erectio Erection E1.0.0.0.0.0.9 Coitus Coitus; Sexual intercourse E1.0.0.0.0.0.10 Ejaculatio1 Ejaculation E1.0.0.0.0.0.11 Emissio Emission E1.0.0.0.0.0.12 Ejaculatio vera Ejaculation proper E1.0.0.0.0.0.13 Semen Semen; Ejaculate E1.0.0.0.0.0.14 Inseminatio Insemination E1.0.0.0.0.0.15 Fertilisatio Fertilization E1.0.0.0.0.0.16 Fecundatio Fecundation; Impregnation E1.0.0.0.0.0.17 Superfecundatio Superfecundation E1.0.0.0.0.0.18 Superimpregnatio Superimpregnation E1.0.0.0.0.0.19 Superfetatio Superfetation E1.0.0.0.0.0.20 Ontogenesis Ontogeny E1.0.0.0.0.0.21 Ontogenesis praenatalis Prenatal ontogeny E1.0.0.0.0.0.22 Tempus praenatale; Tempus gestationis Prenatal period; Gestation period E1.0.0.0.0.0.23 Vita praenatalis Prenatal life E1.0.0.0.0.0.24 Vita intrauterina Intra-uterine life E1.0.0.0.0.0.25 Embryogenesis2 Embryogenesis; Embryogeny E1.0.0.0.0.0.26 Fetogenesis3 Fetogenesis E1.0.0.0.0.0.27 Tempus natale Birth period E1.0.0.0.0.0.28 Ontogenesis postnatalis Postnatal ontogeny E1.0.0.0.0.0.29 Vita postnatalis Postnatal life E1.0.1.0.0.0.1 Mensurae embryonicae et fetales4 Embryonic and fetal measurements E1.0.1.0.0.0.2 Aetas a fecundatione5 Fertilization -

220 Alimentary System: Pancreas
220 Alimentary System: Pancreas Pancreas The exocrine part (C)ispurely serous, and its secretory units, or acini (C15), contain Gross and Microscopic Anatomy polarized epithelial cells. Draining the secre- tory units are long intercalated ducts (C16) The pancreas (A1) is a wedge-shaped organ, that begin within the acini and form the 13–15cm long, that lies on the posterior first part of the excretory duct system. In abdominal wall at the level of L1–L2. It ex- cross-section the invaginated intercalated tends almost horizontally from the C- ducts appear as centroacinar cells (CD17). shaped duodenum to the splenic hilum and The intercalated ducts drain into larger ex- may be divided by its macroscopic features cretory ducts which ultimately unite to form into three parts: the pancreatic duct. The fibrous capsule sur- Head of pancreas (B2). The head of the pan- rounding the pancreas sends delicate creas, which lies in the duodenal loop, is the fibrous septa into the interior of the organ, thickest part of the organ. The hook-shaped dividing it into lobules. uncinate process (B3) projects posteriorly and Alimentary System inferiorly from the head of the pancreas sur- Neurovascular Supply and Lymphatic rounding the mesenteric vessels (B4). Be- Drainage tween the head of the pancreas and the un- Arteries. Arterial supply to the head of the cinate process is a groove called the pan- pancreas, like that of the duodenum (see p. creatic notch (B5). 200), is provided by branches of the ǟ Body of pancreas (B6). Most of the body of gastroduodenal artery ( common hepatic the pancreas lies in front of the vertebral artery): the posterior superior pancreati- column. -

Anatomy of Liver and Spleen
Liver & Spleen 1 Objectives By the end of the lecture, you should be able to describe the: • Location, subdivisions and relations and peritoneal reflection of liver. • Blood supply, nerve supply and lymphatic drainage of liver. • Location, subdivisions and relations and peritoneal reflection of spleen. • Blood supply, nerve supply and lymphatic drainage of spleen. 2 • The largest gland in the body. Liver • Weighs approximately 1500 g. (2.5% of adult body weight). • Lies mainly in: • Right hypochondrium, • Epigastrium and Epigastric • Left hypochondrium. • Protected by the thoracic cage and diaphragm, lies deep to ribs 7-11 on the right side and crosses the midline toward the left nipple. • Moves with the diaphragm and Hypogastric is located more inferiorly in erect posture because of gravity. 3 • Anterior: 1. Diaphragm, Relations of Liver 2. Right and left costal margins, 3. Right and left pleura, 4. Right and left lungs, 5. Xiphoid process, 6. Anterior abdominal wall. • Posterior: 1. Diaphragm, 2. Right kidney, 3. Right suprarenal land, 4. Right colic (hepatic ) flexure, 5. Duodenum, 6. Gallbladder, 7. Inferior vena cava, 8. Esophagus and 9. Fundus of the stomach. 4 Peritoneal Reflection Ø The liver is completely surrounded by a fibrous capsule. Superior Diaphragm layer of Ø It is partially covered by peritoneum. coronary Peritoneum ligament Ø The bare area of the liver is an area lying on the diaphragmatic surface Inferior Anterior layer of abdominal (on posterior surface of right lobe) where there is no coronary wall peritoneum between the liver and the ligament diaphragm. Posterior abdominal Ø Boundaries of Bare area: wall Ø Anterior: Superior layer of coronary ligament. -

Anatomy and Physiology Model Guide Book
Anatomy & Physiology Model Guide Book Last Updated: August 8, 2013 ii Table of Contents Tissues ........................................................................................................................................................... 7 The Bone (Somso QS 61) ........................................................................................................................... 7 Section of Skin (Somso KS 3 & KS4) .......................................................................................................... 8 Model of the Lymphatic System in the Human Body ............................................................................. 11 Bone Structure ........................................................................................................................................ 12 Skeletal System ........................................................................................................................................... 13 The Skull .................................................................................................................................................. 13 Artificial Exploded Human Skull (Somso QS 9)........................................................................................ 14 Skull ......................................................................................................................................................... 15 Auditory Ossicles .................................................................................................................................... -

Anatomical Variation of Ligamentum Teres of Liver - a Case Report
Article ID: WMC003389 ISSN 2046-1690 Anatomical Variation of Ligamentum Teres of Liver - A Case Report Corresponding Author: Mrs. Suni Ebby, Instructor- CCE & CO and Tutor in Anatomy, Department of Anatomy, Gulf Medical University, Ajman, UAE, 4184 - United Arab Emirates Submitting Author: Mrs. Suni Ebby, Instructor- CCE & CO and Tutor in Anatomy, Department of Anatomy, Gulf Medical University, Ajman, UAE, 4184 - United Arab Emirates Article ID: WMC003389 Article Type: Case Report Submitted on:17-May-2012, 02:16:53 PM GMT Published on: 18-May-2012, 05:51:30 PM GMT Article URL: http://www.webmedcentral.com/article_view/3389 Subject Categories:ANATOMY Keywords:Variations, Ligamentum Teres, Quadrate lobe, Liver, Falciform Ligament How to cite the article:Ebby S, Ambike MV. Anatomical Variation of Ligamentum Teres of Liver - A Case Report . WebmedCentral ANATOMY 2012;3(5):WMC003389 Copyright: This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. Source(s) of Funding: None WebmedCentral > Case Report Page 1 of 7 WMC003389 Downloaded from http://www.webmedcentral.com on 18-May-2012, 05:51:30 PM Anatomical Variation of Ligamentum Teres of Liver - A Case Report Author(s): Ebby S, Ambike MV Abstract therapeutic procedures should not be overlooked4. Elmar and Robert stated that after obliteration of umbilical vein over thenext several months after birth, it becomes the ligamentumteres hepatis, which usually Variations related to the liver are frequently measures 10 to 20 cms long5. encountered. This case reports the variant course of ligamentum teres present and its attachment along the Quadrate lobe is distinguished physiologically as the inferior margin of falciform ligament which was medial segment of the left lobe. -
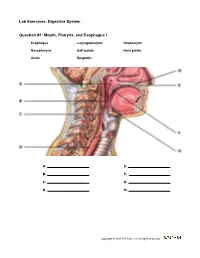
Digestive System
Lab Exercises: Digestive System Question #1: Mouth, Pharynx, and Esophagus I Esophagus Laryngopharynx Oropharynx Nasopharynx Soft palate Hard palate Uvula Epiglottis A. E. B. F. C. G. D. H. Copyright © 2011 A.D.A.M., Inc. All rights reserved. Lab Exercises: Digestive System Question #2: Mouth, Pharynx, and Esophagus II Hyoid bone Thyroid cartilage Cricoid cartilage Trachea A. C. B. D. Copyright © 2011 A.D.A.M., Inc. All rights reserved. Lab Exercises: Digestive System Question # 3: Salivary Glands Parotid gland Parotid duct Sublingual gland Submandibular gland A. C. B. D. Copyright © 2011 A.D.A.M., Inc. All rights reserved. Lab Exercises: Digestive System Question # 4: Small Intestine Pyloric sphincter Duodenum Jejunum Ileum A. C. B. D. Copyright © 2011 A.D.A.M., Inc. All rights reserved. Lab Exercises: Digestive System Question # 5: Large Intestine Cecum Ascending Colon Right (Hepatic) flexure Transverse colon Left (Splenic) flexure Descending colon Sigmoid colon Rectum A. E. B. F. C. G. D. H. Copyright © 2011 A.D.A.M., Inc. All rights reserved. Lab Exercises: Digestive System Question # 6: Frontal Stomach Cardiac part of stomach Gastric rugae Lesser curvature of stomach Pyloric part of stomach Greater curvature of stomach Duodenum Jejunum Pyloric Sphincter A. E. B. F. C. G. D. H. Copyright © 2011 A.D.A.M., Inc. All rights reserved. Lab Exercises: Digestive System Question # 7: Bile Ducts Gallbladder Common hepatic duct Cystic duct Accessory pancreatic duct Common bile duct Main pancreatic duct Hepatopancreatic ampulla Major duodenal papilla A. E. B. F. C. G. D. H. Copyright © 2011 A.D.A.M., Inc. -

Anatomy of Liver and Spleen Doctors Notes Notes/Extra Explanation Please View Our Editing File Before Studying This Lecture to Check for Any Changes
Color Code Important Anatomy of Liver and Spleen Doctors Notes Notes/Extra explanation Please view our Editing File before studying this lecture to check for any changes. Objectives At the end of the lecture, the students should be able to: ✓ Location, subdivisions ,relations and peritoneal reflection of liver. ✓ Blood supply, nerve supply and lymphatic drainage of liver. ✓ Location, subdivisions and relations and peritoneal reflection of spleen. ✓ Blood supply, nerve supply and lymphatic drainage of spleen. Liver o The largest gland in the body. o Weighs approximately 1500 g (approximately 2.5% of adult body weight). o Lies mainly in the right hypochondrium and epigastrium and extends into the left hypochondrium. o Protected by the thoracic cage and diaphragm, its greater part lies deep to ribs 7-11 on the right side and crosses the midline toward the left below the nipple. o Moves with the diaphragm and is located more inferiorly when on is erect (standing) because of gravity. Liver Relations 10:07 Anterior: Extra o Diaphragm o Right & left pleura and lower margins of both lungs o Right and left costal margins o Xiphoid process o Anterior abdominal wall in the subcostal angle Extra Posterior: o Diaphragm o Inferior vena cava o Right kidney and right suprarenal gland o Hepatic flexure of the colon o Duodenum (beginning), gallbladder, esophagus and fundus of the stomach Liver Posterior surface of liver Peritoneal Reflection o The liver is completely surrounded by a fibrous capsule and completely covered by peritoneum (except the bare areas). o The bare area of the liver is a triangular area on the posterior (diaphragmatic) surface of right lobe where there is no intervening peritoneum between the liver and the diaphragm. -

Comparative Anatomical Studies on the Fetal Remnants in Donkey (Asinus Equus) and Camel (Camelus Dromedarius)
IOSR Journal of Agriculture and Veterinary Science (IOSR-JAVS) e-ISSN: 2319-2380, p-ISSN: 2319-2372. Volume 11, Issue 5 Ver. I (May 2018), PP 12-21 www.iosrjournals.org Comparative Anatomical Studies on the Fetal Remnants in Donkey (Asinus Equus) And Camel (Camelus Dromedarius) Nora, A. shaker and Samar M. EL Gammal Department of Anatomy and Embryology, Faculty of Veterinary Medicine, Cairo University Abstract Aim: The present study described the anatomical structures of the umbilical cord, fetal circulation and their remnants in the adult. This work will help veterinary practitioners in diagnosis of the abnormalities of these umbilical structures in newly born foals and camel calves. Material and methods: it was applied on five specimens from donkeys and camels; three aborted feti and two apparently healthy adults. Fetal specimens were injected with the colored latex in the umbilical veins and arteries then preserved in a 10% formalin solution for two days before dissection. The structures of the umbilical cord and the embryonic structures were identified and photographed. The adult specimens were treated by ordinary routine method of preservation; mixture of 10%formalin, 4%phenol and 1% glycerin for two days then dissected to identify the fetal remnants. Result: The umbilical cord contained four structures in donkey; single umbilical vein, two umbilical arteries and the urachus, while in camel, it contained the same structures but two umbilical veins instead of one. In the adult, these fetal tubes were occluded as the round ligament of liver, round ligaments of urinary bladder and cicatrix on the bladder apex. The other fetal structures were; the foramen ovale and the ductus arteriosus and the paired gubernaculua related to the testes in males and the ovaries in females.