A Taxonomic Review of the Yellow-Flowered Species Of
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Rhododendron Ferrugineum L
Volume 18(1), 123- 130, 2014 JOURNAL of Horticulture, Forestry and Biotechnology www.journal-hfb.usab-tm.ro Rhododendron ferrugineum L. and Rhododendron myrtifolium Schott & Kotschy in habitats from Eastern Alps mountains and Carpathian Mountains Căprar M.1,2, Cantor Maria2*, Szatmari P.1, Sicora C.1 1)Biological Research Center, Botanical Garden ,,Vasile Fati” Jibou, Parcului Street,no.14,455200 Jibou, Romania; 2)University of Agricultural Sciences and Veterinary Medicine Cluj-Napoca, Faculty of Horticulture, Mănăștur Street, no 3-5,4000472 Cluj-Napoca, Romania; * Corresponding author. Email: [email protected] Abstract This paper presents results of research carried on two species Key words of Rhododendron in habitats from different regions of Central and Eastern Europe (Rhododendron ferrugineum and Rhododendron myrtifolium). It rhododendrons species, presents the ecological requirements of each habitat, their spread, main plant habitats, plant communities, association and floristic composition based on the dominance of probative Carpathian Mountains, Alps species. A correlation is made between habitats from different classifications, Mountains but with the same features, mentioning EUNIS codes, Emerald, Natura 2000, Palaearctic Habitats and the European forest types. This paper presents information on the spread of two types of habitats containing Rhododendronfrom Europe, the environmental conditions in which they live and the accompanying species involved, more or less, in the composition of habitats. It describes the types of vegetation in the Alps (Austria) and the Carpathian Mountains (Romania). Vegetation was observed following the research in the field. Knowledge of the habitats in which of Australia, with over 200 species only in the island of Rhododendron species live becomes an important New Guinea [2]. -

Characterizing the Genetic Variation in Seven Species of Deciduous Native Azaleas and Identifying the Mechanism of Azalea Lacebug Resistance in Deciduous Azalea
CHARACTERIZING THE GENETIC VARIATION IN SEVEN SPECIES OF DECIDUOUS NATIVE AZALEAS AND IDENTIFYING THE MECHANISM OF AZALEA LACEBUG RESISTANCE IN DECIDUOUS AZALEA by MATTHEW RANDOLPH CHAPPELL (Under the direction of Dr. Carol Robacker) ABSTRACT Despite the ecologic and economic importance of native deciduous azaleas (Rhododendron spp. section Pentanthera), our understanding of interspecific variation of North American deciduous azalea species is limited. Furthermore, little is known concerning intraspecific or interpopulation genetic variation. The present study addresses questions of genetic diversity through the use of amplified fragment length polymorphism (AFLP) analysis. Twenty-five populations of seven species of native azalea were analyzed using three primer pairs that amplified a total of 417 bands. Based on analysis of molecular variance (AMOVA) and estimates of Nei’s coefficients of gene diversity (HS, HT, and GST), the majority of variation in deciduous azalea occurs within populations. Both among species and among population variation was low, likely the effect of common ancestry as well as frequent introgression among members (and populations) of section Pentanthera. The majority of populations were grouped into species based on Nei’s unbiased genetic distances viewed as a UPGMA phenogram. The significance of these results is discussed in relation to breeding in section Pentanthera. In addition to the lack of information concerning genetic variation in North American native azaleas, little is known concerning the insect-plant interaction between the primary azalea pest in the United States, azalea lace bug (ALB) (Stephanitis pyrioides Scott), and deciduous azalea. Azaleas are largely resistant to predation by insects, with the exception of ALB. Within deciduous azalea (Rhododendron section Pentanthera) varying levels of resistance to ALB is observed with a continuous distribution from susceptible to highly resistant. -

The Red List of Rhododendrons
The Red List of Rhododendrons Douglas Gibbs, David Chamberlain and George Argent BOTANIC GARDENS CONSERVATION INTERNATIONAL (BGCI) is a membership organization linking botanic gardens in over 100 countries in a shared commitment to biodiversity conservation, sustainable use and environmental education. BGCI aims to mobilize botanic gardens and work with partners to secure plant diversity for the well-being of people and the planet. BGCI provides the Secretariat for the IUCN/SSC Global Tree Specialist Group. Published by Botanic Gardens Conservation FAUNA & FLORA INTERNATIONAL (FFI) , founded in 1903 and the International, Richmond, UK world’s oldest international conservation organization, acts to conserve © 2011 Botanic Gardens Conservation International threatened species and ecosystems worldwide, choosing solutions that are sustainable, are based on sound science and take account of ISBN: 978-1-905164-35-6 human needs. Reproduction of any part of the publication for educational, conservation and other non-profit purposes is authorized without prior permission from the copyright holder, provided that the source is fully acknowledged. Reproduction for resale or other commercial purposes is prohibited without prior written permission from the copyright holder. THE GLOBAL TREES CAMPAIGN is undertaken through a partnership between FFI and BGCI, working with a wide range of other The designation of geographical entities in this document and the presentation of the material do not organizations around the world, to save the world’s most threatened trees imply any expression on the part of the authors and the habitats in which they grow through the provision of information, or Botanic Gardens Conservation International delivery of conservation action and support for sustainable use. -

THE POTENTIAL WILDERNESS AREA GROSSVENEDIGER a Report to the Wild Europe Initiative
IN COOPERATION WITH THE POTENTIAL WILDERNESS AREA GROSSVENEDIGER A report to the Wild Europe Initiative The Potential Wilderness Area Grossvenediger, page 1 THE POTENTIAL WILDERNESS AREA GROSSVENEDIGER A report to the Wild Europe Initiative published by: WWF Austria, 2014 written by: • Bernhard Kohler, Head of Austrian programme, WWF Austria, Ottakringerstr. 114-16, 1160 Vienna, Austria, [email protected] • Michael Zika, Wilderness officer, WWF Austria, Ottakringerstr. 114-16, 1160 Wien, Vienna, Austria, [email protected] • Vlado Vancura, Director Wilderness Development, European Wilderness Society, Dechant Franz Fuchs Str., 5580 Tamsweg, Austria, [email protected] Citation: Kohler, B., M. Zika & V. Vancura (2014): The potential wilderness area Grossvenediger. A report to the Wild Europe Initiative. WWF Austria, Vienna, 92 pp. Download from: http://www.wwf.at/de/wildnis-downloads/ Acknowledgements: The present study was produced by WWF Austria, on behalf of National Park Hohe Tauern Salzburg, with support from the European Wilderness Society. Maps and various data have been kindly provided by the National Park administration. Our thanks go to director Wolfgang Urban, vice-director Ferdinand Lainer, Barbara Hochwimmer and Kristina Bauch. LIST OF FIGURES Fig.1. The visiting group (from left to right): Bernhard Kohler and Michael Zika (WWF Austria), Vlado Vancura (European Wilderness Society), Wolfgang Urban (NP Director) .......................................................................................16 INHALT Fig.2. Zonation of Hohe Tauern national park: core zones in dark green, external zones in light green, special protection areas in yellow. Provincial borders of Tyrol, Salzburg and Carinthia shown as dotted red lines. ......................................18 Fig.3. Sign announcing core zone ...............................................................................................................................................22 Fig.4. Delimitation and zonation of the proposed wilderness area. -

The Hills Are Alive
RHS LEVEL 4 DIPLOMA IN HORTICULTURAL PRACTICE MOUNTAIN PLANTS OF THE EUROPEAN ALPS THE HILLS ARE ALIVE CAN GARDENS SHOWCASE AND BETTER EXPLAIN THE DIVERSITY OF MOUNTAIN PLANTS FOUND IN THE EUROPEAN ALPS AND THE THREATS THAT THEY FACE BY DISPLAYING PLANTS IN PLANT COMMUNITIES? DANIEL JONES RESEARCH PROJECT 2019 - 2020 MOUNTAIN PLANTS OF THE EUROPEAN ALPS RESEARCH PROJECT 2019 - 2020 ACKNOWLEDGEMENTS THE AUTHOR WISHES TO THANK Caroline Jackson for her guidance in writing this research project. The alpine horticultural teams at RHS Garden Wisley and the Royal Botanic Gardens, Kew for their encouragement, guidance and expertise in the cultivation of alpine plants. The gardeners of the Schynige Platte Alpine Garden for passing on their knowledge and passion for alpine flora and for inspiring me to undertake this research project. Adrian Möhl for passing on his knowledge and expertise of the wild plants of Switzerland. The visitors of RHS Garden Wisley for answering my questionnaire as part of this research project. The scientists and wardens of the Swiss National Park for passing on their knowledge of wild plant communities, biodiversity in the Alps and its conservation. The gardeners of the University of Basel, University of Zürich and Juragarten for their knowledge and expertise in the cultivation of Switzerland’s flora. Dr Elinor Breman and the partners of the Alpine Seed Conservation & Research Network for encouraging me to understand more about the flora of the Alps and its conservation. Dawn Rushen for her help in proofreading this project. DANIEL JONES PAGE 2 2020 MOUNTAIN PLANTS OF THE EUROPEAN ALPS RESEARCH PROJECT 2019 - 2020 ABSTRACT There are over 900 plant communities in the Alps and this research project explores how these can be used in UK gardens. -

Biological Activities and Cytotoxicity of Leaf Extracts from Plants of the Genus Rhododendron
From Ethnomedicine to Application: Biological Activities and Cytotoxicity of Leaf Extracts from Plants of the Genus Rhododendron by Ahmed Rezk a Thesis submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in Biochemistry Approved Dissertation Committee Prof. Dr. Matthias Ullrich, Prof. of Microbiology Prof. Dr. Klaudia Brix, Prof. of Cell Biology Jacobs University Bremen Prof. Dr. Nikolai Kuhnert Prof. of Chemistry Jacobs University Bremen Prof. Dr. Dirk Albach, Prof. of Plant Biodiversity University of Oldenburg Date of Defense: 15.06.2015 This PhD thesis project was financed by Stiftung Rhododendronpark Bremen Dedicated to: My Wife Rasha Acknowledgment Acknowledgment First, I thank Allah for giving me the ability and strength to accomplish this study. I would like to express my gratitude to the following people for support during my work: I would like to express my sincere appreciation and gratitude to my PhD supervisors, Prof. Dr. Matthias Ullrich, and Prof. Dr. Klaudia Brix, who gave me the opportunity to compose my doctoral thesis in their workgroups. I would like to thank them for their support, guidance and all the time they gave to discuss and help in designing experiments to achieve this work. I would also like to thank my dissertation committee members, Prof. Dr. Nikolai Kuhnert and Prof. Dr. Dirk Albach for their time and for their valuable comments during our meetings and reviewing my thesis. I would specifically like to thank AG Ullrich and AG Brix lab members, Amna Mehmood, Antje Stahl, Gabriela Alfaro-Espinoza, Khaled Abdallah, Neha Kumari, Maria Qatato, Joanna Szumska, and Jonas Weber for maintaining a friendly and family working environment. -

Alpine Flowers of the Swiss Alps
Wengen - Alpine Flowers of the Swiss Alps Naturetrek Tour Report 17 - 24 June 2018 Antennaria dioica Crocus vernus Lauterbrunnen valley Pulsatilla vernalis Report and images by David Tattersfield Naturetrek Mingledown Barn Wolf's Lane Chawton Alton Hampshire GU34 3HJ UK T: +44 (0)1962 733051 E: [email protected] W: www.naturetrek.co.uk Tour Report Wengen - Alpine Flowers of the Swiss Alps Tour participants: David Tattersfield (tour leader) with 16 Naturetrek clients. Day 1 Sunday 17th June We arrived in Zurich in the early afternoon and took the comfortable inter-city trains to Interlaken. Here we joined the regional train and followed the milky waters of the Lutschine River to Lauterbrunnen. The last leg of the journey was on the cogwheel railway to Wengen, where it was just a short walk to our hotel. We arrived around 5.30pm, with time to settle in before dinner. Day 2 Monday 18th June We enjoyed a sunny day with cloud drifting over the mountain tops and only thickening during the late afternoon. We took the Mannlichen cable-car, high above the village and spent the morning exploring a wide range of habitats, as we made our way towards the summit, at 2343 metres. Inevitably, we made slow progress, with new discoveries every few paces. The mountain pasture was speckled with the white flowers of Kupfer’s Buttercup Ranunculus kuepferi and drifts of Globeflower Trollius europaeus made rich-yellow patches on the cliff- tops. Here and there were colourful patches of Spring Gentian Gentiana verna, Bird’s-eye Primrose Primula farinosa and Whorled Lousewort Pedicularis verticillata and near the edges of melting snowdrifts, we found Oxlip Primula elatior, the delightful frilled flowers of Alpine Snowbell Soldanella alpina and patches of Spring Crocus Crocus vernus. -

Pollen Morphology and Its Systematic Significance in the Ericaceae
Title Pollen Morphology and Its Systematic Significance in the Ericaceae Author(s) Sawara, A.K.M. Golam Citation 北海道大学. 博士(農学) 甲第8187号 Issue Date 2007-03-23 DOI 10.14943/doctoral.k8187 Doc URL http://hdl.handle.net/2115/46925 Type theses (doctoral) File Information sarwar.pdf Instructions for use Hokkaido University Collection of Scholarly and Academic Papers : HUSCAP Pollen Morphology and Its Systematic Significance in the Ericaceae (ツツジ科植物の花粉形態とその体系学的意義) A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy By Sarwar, A.K.M. Golam Division of Bioresources and Product Science Graduate School of Agriculture Hokkaido University Sapporo, Japan March, 2007 Contents Abstract iv Chapter 1: GENERAL INTRODUCTION 1 Chapter 2: MATERIALS AND METHODS 10 Chapter 3: POLLEN MORPHOLOGY AND ITS SYSTEMATIC SIGNIFICANCE 20 GENERAL POLLEN MORPHOLOGY OF THE ERICACEAE 20 3-1 SUBFAMILY ENKIANTHOIDEAE 24 Introduction 24 Results 25 Discussion 30 3-2 SUBFAMILY ARBUTOIDEAE 44 Introduction 44 Results 45 Discussion 51 3-3 SUBFAMILY ERICOIDEAE 60 Introduction 60 Results 61 Discussion 81 3-4 SUBFAMILY CASSIOPOIDEAE 106 Introduction 106 Results 107 Discussion 110 ii 3-5 SUBFAMILY HARRIMANELLOIDEAE 112 Introduction 112 Results 113 Discussion 113 3-6 SUBFAMILY VACCINIOIDEAE 118 Introduction 118 Results 119 Discussion 160 Chapter 4: GENERAL DISCUSSION 203 Acknowledgements 252 Summary 254 References 259 Appendix I: Different classification systems of Ericaceae 281 Appendix II: Specimens examined 287 iii Abstract A detailed description of the range of pollen morphological variation within the family Ericaceae sensu Kron et al. (2002a) has been presented. For this palynological investigation, 275 taxa of 270 species representing 57 genera and 6 subfamilies were studied with light (LM) and scanning electron microscopy (SEM), and 31 species with transmission electron microscopy (TEM). -

Molecular Phylogenetics and Patterns of Floral Evolution in the Ericales
Int. J. Plant Sci. 166(2):265–288. 2005. Ó 2005 by The University of Chicago. All rights reserved. 1058-5893/2005/16602-0009$15.00 MOLECULAR PHYLOGENETICS AND PATTERNS OF FLORAL EVOLUTION IN THE ERICALES Ju¨rg Scho¨nenberger,1,* Arne A. Anderberg,y and Kenneth J. Sytsma* *Department of Botany, University of Wisconsin, Madison, Wisconsin 53706-1831, U.S.A.; and yDepartment of Phanerogamic Botany, Swedish Museum of Natural History, SE-104 05 Stockholm, Sweden The diverse and species-rich order Ericales has found considerable interest among systematists in recent years. Molecular phylogenetic studies not only have convincingly demonstrated the monophyly of the order, comprising 23 families formerly placed in three different subclasses (Asteridae, Dilleniidae, and Rosidae), but have also resolved Ericales as sister to euasterids. Most ericalean families are well circumscribed and have been or are currently subject to intrafamilial phylogenetic studies. In spite of all the attention that Ericales have received recently, there remains a major challenge, the still largely unresolved deeper nodes in the ericalean phylogeny. This study aims to improve our current knowledge of the interfamilial relationships by expanding on gene and taxon sampling and to evaluate the evolution of important floral characters in light of the resulting phylogeny. We add a nuclear region (26s rDNA) to already published data sets (nuclear: 18s rDNA; mitochondrial: atp1, matR; chloroplast: atpB, ndhF, rbcL, matK, the rps16 intron, the trnT-trnF spacer, and the trnV-atpE spacer), for a total of 11 molecular markers that include nearly 20 kb of sequences. Our analyses, applying both maximum parsimony and Bayesian inference, resolve some of the deeper nodes in the phylogeny. -
Could Bryophagous Beetles (Coleoptera: Byrrhidae) Help Us Understand Bryophyte Taxonomy? Preferences Within the Hypnum Cupressiforme Hedw
plants Article Could Bryophagous Beetles (Coleoptera: Byrrhidae) Help Us Understand Bryophyte Taxonomy? Preferences within the Hypnum cupressiforme Hedw. Species Complex Petr Pyszko 1,*, Michaela Drgová 1, Stanislav Ožana 1, OndˇrejDor ˇnák 1, David Rožek 2, Daniel Lee Cˇ íp 2, VítˇezslavPlášek 1 and Pavel Drozd 1 1 Department of Biology and Ecology/Institute of Environmental Technologies, Faculty of Science, University of Ostrava, Chittussiho 10, 710 00 Ostrava, Czech Republic; [email protected] (M.D.); [email protected] (S.O.); [email protected] (O.D.); [email protected] (V.P.); [email protected] (P.D.) 2 Heyrovsky High School of Chemistry, Stˇredoškolská 2854/1, 700 30 Ostrava, Czech Republic; [email protected] (D.R.); [email protected] (D.L.C.)ˇ * Correspondence: [email protected]; Tel.: +420-608-546-079 Abstract: Intrataxonomic differences in terms of angiosperm suitability for herbivorous insects stem from variables such as plant structure, palatability, and chemistry. It has not yet been elucidated whether these differences also occur in terms of the bryophyte’s suitability to bryophages. Hypnum cupressiforme Hedw. is a morphologically variable moss species frequently inhabited or fed by insects. In this investigation, we offered five morphotypes of H. cupressiforme to two bryophagous species of Citation: Pyszko, P.; Drgová, M.; Byrrhidae (Coleoptera) to reveal whether the intrataxonomic variability affects beetles’ preferences. Ožana, S.; Dorˇnák,O.; Rožek, D.; Lee The morphotypes were offered with preserved and removed spatial structures. There were no Cíp,ˇ D.; Plášek, V.; Drozd, P. Could significant differences in morphotype preferences when spatial structures were preserved, although Bryophagous Beetles (Coleoptera: flat usual turgid Byrrhidae) Help Us Understand during the daytime, the beetles moved from the morphotype to the and morphotypes. -
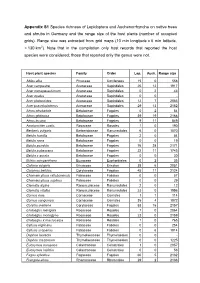
Appendix S1 Species Richness of Lepidoptera and Auchenorrhyncha on Native Trees and Shrubs in Germany and the Range Size of the Host Plants (Number of Occupied Grids)
Appendix S1 Species richness of Lepidoptera and Auchenorrhyncha on native trees and shrubs in Germany and the range size of the host plants (number of occupied grids). Range size was extracted from grid maps (10 min longitude x 6 min latitude, ≈ 130 km 2). Note that in the compilation only host records that reported the host species were considered; those that reported only the genus were not. Host plant species Family Order Lep. Auch. Range size Abies alba Pinaceae Coniferales 15 0 558 Acer campestre Aceraceae Sapindales 25 12 1917 Acer monspessulanum Aceraceae Sapindales 0 2 43 Acer opalus Aceraceae Sapindales 0 0 - Acer platanoides Aceraceae Sapindales 12 7 2083 Acer pseudoplatanus Aceraceae Sapindales 29 13 2152 Alnus alnobetula Betulaceae Fagales 0 2 54 Alnus glutinosa Betulaceae Fagales 39 19 2168 Alnus incana Betulaceae Fagales 9 11 849 Amelanchier ovalis Rosaceae Rosales 1 0 190 Berberis vulgaris Berberidaceae Ranunculales 6 0 1070 Betula humilis Betulaceae Fagales 2 0 54 Betula nana Betulaceae Fagales 0 0 19 Betula pendula Betulaceae Fagales 76 28 2171 Betula pubescens Betulaceae Fagales 22 11 1745 Betula x aurata Betulaceae Fagales 0 0 30 Buxus sempervirens Buxaceae Euphorbiales 0 2 35 Calluna vulgaris Ericaceae Ericales 38 6 2051 Carpinus betulus Corylaceae Fagales 45 11 2124 Chamaecytisus ratisbonensis Fabaceae Fabales 0 0 57 Chamaecytisus supinus Fabaceae Fabales 0 0 29 Clematis alpina Ranunculaceae Ranunculales 2 0 12 Clematis vitalba Ranunculaceae Ranunculales 23 0 1556 Cornus mas Cornaceae Cornales 1 1 114 Cornus sanguinea -

Disparity in Sr and Cs Uptake in Alpine Plants
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by RERO DOC Digital Library Plant Soil (2012) 355:29–39 DOI 10.1007/s11104-011-1110-6 REGULAR ARTICLE Disparity in 90Sr and 137Cs uptake in Alpine plants: phylogenetic effect and Ca and K availability Thomas Guillaume & Fabienne Chawla & Philipp Steinmann & Jean-Michel Gobat & Pascal Froidevaux Received: 8 September 2011 /Accepted: 15 December 2011 /Published online: 4 February 2012 # Springer Science+Business Media B.V. 2012 Abstract on 6 undisturbed and geochemically different soils in Background and aims Uptake of 90Sr and 137Cs in the Alpine valley of Piora, Switzerland. plants varies widely between soil types and between Results Results show that a strong correlation exists plant species. It is now recognized that the radionu- between the log TF and the log of exchangeable Ca or clide uptake in plants is more influenced by site- K of the soils. specific and plant-specific parameters rather than the Conclusions Cs uptake by Phleum rhaeticum (Poales) bulk radionuclide concentration in soil. We hypothe- and Alchemilla xanthochlora (Rosales) is more sensi- sized that the stress which Alpine plants experience tive to the amount of exchangeable K in the soil than because of the short growing season may enhance the the corresponding uptake by other orders. Moreover, phylogenetic effect on the 137Cs and 90Sr transfer the 90Sr results indicate a phylogenetic effect between factors as well as the dependency of the uptake by Non-Eudicot and Eudicots: the order Poales (Phleum plant to the concentrations of exchangeable Ca and K rhaeticum) concentrating much less 90Sr than Eudicots of soils.