Related Kinases (VRK) Bound to Small-Molecule Inhibitors Identifies Different P-Loop Conformations
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Fig. L COMPOSITIONS and METHODS to INHIBIT STEM CELL and PROGENITOR CELL BINDING to LYMPHOID TISSUE and for REGENERATING GERMINAL CENTERS in LYMPHATIC TISSUES
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date Χ 23 February 2012 (23.02.2012) WO 2U12/U24519ft ft A2 (51) International Patent Classification: AO, AT, AU, AZ, BA, BB, BG, BH, BR, BW, BY, BZ, A61K 31/00 (2006.01) CA, CH, CL, CN, CO, CR, CU, CZ, DE, DK, DM, DO, DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, (21) International Application Number: HN, HR, HU, ID, IL, IN, IS, JP, KE, KG, KM, KN, KP, PCT/US201 1/048297 KR, KZ, LA, LC, LK, LR, LS, LT, LU, LY, MA, MD, (22) International Filing Date: ME, MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, 18 August 201 1 (18.08.201 1) NO, NZ, OM, PE, PG, PH, PL, PT, QA, RO, RS, RU, SC, SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, (25) Filing Language: English TN, TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, (26) Publication Language: English ZW. (30) Priority Data: (84) Designated States (unless otherwise indicated, for every 61/374,943 18 August 2010 (18.08.2010) US kind of regional protection available): ARIPO (BW, GH, 61/441,485 10 February 201 1 (10.02.201 1) US GM, KE, LR, LS, MW, MZ, NA, SD, SL, SZ, TZ, UG, 61/449,372 4 March 201 1 (04.03.201 1) US ZM, ZW), Eurasian (AM, AZ, BY, KG, KZ, MD, RU, TJ, TM), European (AL, AT, BE, BG, CH, CY, CZ, DE, DK, (72) Inventor; and EE, ES, FI, FR, GB, GR, HR, HU, IE, IS, ΓΓ, LT, LU, (71) Applicant : DEISHER, Theresa [US/US]; 1420 Fifth LV, MC, MK, MT, NL, NO, PL, PT, RO, RS, SE, SI, SK, Avenue, Seattle, WA 98101 (US). -

WO 2018/223101 Al 06 December 2018 (06.12.2018) W !P O PCT
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date WO 2018/223101 Al 06 December 2018 (06.12.2018) W !P O PCT (51) International Patent Classification: (71) Applicant: JUNO THERAPEUTICS, INC. [US/US]; 400 A 61K 35/1 7 (20 15.0 1) A 61P 35/00 (2006 .0 1) Dexter Ave. N., Suite 1200, Seattle, WA 98109 (US). (21) International Application Number: (72) Inventor: ALBERTSON, Tina; 400 Dexter Ave. N., Suite PCT/US2018/035755 1200, Seattle, WA 98109 (US). (22) International Filing Date: (74) Agent: AHN, Sejin et al; Morrison & Foerster LLP, 1253 1 0 1 June 2018 (01 .06.2018) High Bluff Drive, Suite 100, San Diego, CA 92130-2040 (US). (25) Filing Language: English (81) Designated States (unless otherwise indicated, for every (26) Publication Language: English kind of national protection available): AE, AG, AL, AM, (30) Priority Data: AO, AT, AU, AZ, BA, BB, BG, BH, BN, BR, BW, BY, BZ, 62/5 14,774 02 June 2017 (02.06.2017) US CA, CH, CL, CN, CO, CR, CU, CZ, DE, DJ, DK, DM, DO, 62/5 15,530 05 June 2017 (05.06.2017) US DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, HN, 62/521,366 16 June 2017 (16.06.2017) u s HR, HU, ID, IL, IN, IR, IS, JO, JP, KE, KG, KH, KN, KP, 62/527,000 29 June 2017 (29.06.2017) u s KR, KW, KZ, LA, LC, LK, LR, LS, LU, LY, MA, MD, ME, 62/549,938 24 August 2017 (24.08.2017) u s MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, 62/580,425 0 1 November 2017 (01 .11.2017) u s OM, PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SA, 62/593,871 0 1 December 2017 (01 .12.2017) u s SC, SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, TN, 62/596,764 08 December 2017 (08.12.2017) u s TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW. -

Nick-Thomas.Pdf
Innovating Pre-Clinical Drug Development Towards an Integrated Approach to Investigative Toxicology in Human Cell Models Nick Thomas PhD Principal Scientist Cell Technologies GE Healthcare ELRIG Pharmaceutical Flow Cytometry & Imaging 2012 10-11th October 2012, AstraZeneca, Alderley Park Drug Toxicology Current issues – problems & solutions Using animal models to reflect Quality and robustness of Testing multiple endpoints human responses toxicity cell models leading to false-positives • animals ≠ humans • scarcity of primary cells/tissues • multiple testing increases • animal ≠ animal • source variability sensitivity at cost of specificity • cross species testing may • more abundant models • different assay combinations increase sensitivity but (immortalized/genetically yield varying predictivity decrease specificity engineered cells) may have • testing multiple endpoints leads • metabolism & MOA ? reduced predictivity to false positives Sensitivity Specificity Assay Combinations Integrate range of predictive Integrate robust human stem Integrate and standardize human cell models cell derived models most predictive parameters Cytiva™ Cardiomyocytes H7 hESC Cardiomyocytes 109 Expansion Differentiation 0 5 10 15 20 25 30 Media 1 Media 2 Growth Factors Feed Cytiva Cardiomyocytes DNA Troponin I DNA Connexin 43 Troponin I HCA Biochemical Assays Patch Clamp Impedance Multi-Electrode Arrays Respiration HCA Biochemical Assays Patch Clamp Impedance Multi-Electrode Arrays Respiration Cardiotoxicity Profiling of Anticancer Drugs Cytiva Cardiomyocytes & IN Cell Analyzer Cardiotoxicity & Anticancer Drugs Drug Pipelines Toxicity in Drug Development Hepatotoxicity Nephrotoxicity Cardiotoxicity Rhabdomyolysis Other Data from: Drug pipeline: Q411. Mak H.C. 2012 Data from; Wilke RA et al. Nature Reviews Drug Discovery Nature Biotechnol. 30,15 2007 6, 904-916 Cardiotoxicity of Anticancer Drugs Off Target On Target : Off Therapy DOX GATA4 ROS Bcl2 Cell Death Adapted from; Kobayashi S. -

DRUG and MEDICAL DEVICE HIGHLIGHTS Helping You Maintain and Improve Your Health 2019
DRUG AND MEDICAL DEVICE HIGHLIGHTS Helping you maintain and improve your health 2019 DRUG AND MEDICAL DEVICE HIGHLIGHTS 2019 Helping you maintain and improve your health Learn about the new drugs and medical devices that Health Canada approved for sale in Canada, the information we published about potential safety issues, and our other accomplishments in 2019. Health Canada is the federal department responsible for helping the people of Canada maintain and improve their health. Health Canada is committed to improving the lives of all of Canada’s people and to making this country’s population among the healthiest in the world as measured by longevity, lifestyle and effective use of the public health care system. Également disponible en français sous le titre : Préserver et améliorer votre santé : Faits saillants sur les médicaments et les instruments médicaux 2019 To obtain additional information, please contact: Health Canada Address Locator 0900C2 Ottawa, ON K1A 0K9 Tel.: 613-957-2991 Toll free: 1-866-225-0709 Fax: 613-941-5366 TTY: 1-800-465-7735 E-mail: [email protected] © Her Majesty the Queen in Right of Canada, as represented by the Minister of Health, 2020 Publication date: May 2020 This publication may be reproduced for personal or internal use only without permission provided the source is fully acknowledged. Cat.: H161-11E-PDF ISSN: 2562-9816 Pub.: 190484 CONTENTS WELCOME TO OUR 2019 HIGHLIGHTS REPORT ..........................................................................................1 MESSAGE FROM THE CHIEF MEDICAL -

Two Cases of Immediate Stent Fracture After Zotarolimus-Eluting
Case Report http://dx.doi.org/10.4070/kcj.2015.45.1.67 Print ISSN 1738-5520 • On-line ISSN 1738-5555 Korean Circulation Journal Two Cases of Immediate Stent Fracture after Zotarolimus-Eluting Stent Implantation Pil Hyung Lee, MD, Seung-Whan Lee, MD, Jong-Young Lee, MD, Young-Hak Kim, MD, Cheol Whan Lee, MD, Duk-Woo Park, MD, Seong-Wook Park, MD, and Seung-Jung Park, MD Department of Cardiology, University of Ulsan College of Medicine, Asan Medical Center, Seoul, Korea Drug-eluting stent (DES) implantation is currently the standard treatment for various types of coronary artery disease. However, previous reports indicate that stent fractures, which usually occur after a period of time from the initial DES implantation, have increased during the DES era; stent fractures can contribute to unfavorable events such as in-stent restenosis and stent thrombosis. In our present report, we describe two cases of zotarolimus-eluting stent fracture: one that was detected six hours after implementation, and the other case that was detected immediately after deployment. Both anatomical and technical risk factors contributed to these unusual cases of imme- diate stent fracture. (Korean Circ J 2015;45(1):67-70) KEY WORDS: Drug-eluting stents; Percutaneous coronary intervention; Complications. Introduction eluting stent (ZES). Stent fractures have recently become an important concern in Cases the medical community due to their potential association with se- rious conditions such as in-stent restenosis (ISR) and stent throm- Case 1 bosis after drug-eluting stent (DES) implantation.1) Most currently A 62-year-old man, who had undergone cardiac transplantation reported stent fractures are found in lesions implanted with siroli- for advanced heart failure, was referred to our catheterization labo- mus-eluting stents (SES), likely due to the inherent platform material ratory for a regular surveillance coronary angiogram and concomi- and design of such stents.2) Furthermore, while the exact timing of tant intravascular ultrasound (IVUS) imaging. -

Physiologically Based Pharmacokinetic Modeling in Regulatory Decision‐Making at the European Medicines Agency
ARTICLES Physiologically Based Pharmacokinetic Modeling in Regulatory Decision-Making at the European Medicines Agency E Luzon1, K Blake1, S Cole2,3,4, A Nordmark4,5, C Versantvoort3,6 and E Gil Berglund3,5 Physiologically based pharmacokinetic (PBPK) modeling is a valuable tool in drug development and regulatory assessment, as it offers the opportunity to simulate the pharmacokinetics of a compound, with a mechanistic understanding, in a variety of populations and situations. This work reviews the use and impact of such modeling in selected regulatory procedures submitted to the European Medicines Agency (EMA) before the end of 2015, together with its subsequent reflection in public documents relating to the assessment of these procedures. It is apparent that the reference to PBPK modeling in regulatory public documents underrepresents its use. A positive trend over time of the number of PBPK models submitted is shown, and in a number of cases the results of these may impact the decision- making process or lead to recommendations in the product labeling. These results confirm the need for regulatory guidance in this field, which is currently under development by the EMA. Study Highlights WHAT IS THE CURRENT KNOWLEDGE ON THE WHAT THIS STUDY ADDS TO OUR KNOWLEDGE TOPIC? þ This is the first work presenting the situation in the Europe- þ PBPK is a cutting-edge technology that is increasingly being an Union and confirms what was detected by the FDA is a applied to drug development to address various aspects related global trend. to clinical pharmacology. Previous works by the FDA acknowl- HOW THIS MIGHT CHANGE CLINICAL PHARMA- edge its use in regulatory submissions in the US, but so far there COLOGY OR TRANSLATIONAL SCIENCE is no analysis of the European perspective. -

Drug Name Plate Number Well Location % Inhibition, Screen Axitinib 1 1 20 Gefitinib (ZD1839) 1 2 70 Sorafenib Tosylate 1 3 21 Cr
Drug Name Plate Number Well Location % Inhibition, Screen Axitinib 1 1 20 Gefitinib (ZD1839) 1 2 70 Sorafenib Tosylate 1 3 21 Crizotinib (PF-02341066) 1 4 55 Docetaxel 1 5 98 Anastrozole 1 6 25 Cladribine 1 7 23 Methotrexate 1 8 -187 Letrozole 1 9 65 Entecavir Hydrate 1 10 48 Roxadustat (FG-4592) 1 11 19 Imatinib Mesylate (STI571) 1 12 0 Sunitinib Malate 1 13 34 Vismodegib (GDC-0449) 1 14 64 Paclitaxel 1 15 89 Aprepitant 1 16 94 Decitabine 1 17 -79 Bendamustine HCl 1 18 19 Temozolomide 1 19 -111 Nepafenac 1 20 24 Nintedanib (BIBF 1120) 1 21 -43 Lapatinib (GW-572016) Ditosylate 1 22 88 Temsirolimus (CCI-779, NSC 683864) 1 23 96 Belinostat (PXD101) 1 24 46 Capecitabine 1 25 19 Bicalutamide 1 26 83 Dutasteride 1 27 68 Epirubicin HCl 1 28 -59 Tamoxifen 1 29 30 Rufinamide 1 30 96 Afatinib (BIBW2992) 1 31 -54 Lenalidomide (CC-5013) 1 32 19 Vorinostat (SAHA, MK0683) 1 33 38 Rucaparib (AG-014699,PF-01367338) phosphate1 34 14 Lenvatinib (E7080) 1 35 80 Fulvestrant 1 36 76 Melatonin 1 37 15 Etoposide 1 38 -69 Vincristine sulfate 1 39 61 Posaconazole 1 40 97 Bortezomib (PS-341) 1 41 71 Panobinostat (LBH589) 1 42 41 Entinostat (MS-275) 1 43 26 Cabozantinib (XL184, BMS-907351) 1 44 79 Valproic acid sodium salt (Sodium valproate) 1 45 7 Raltitrexed 1 46 39 Bisoprolol fumarate 1 47 -23 Raloxifene HCl 1 48 97 Agomelatine 1 49 35 Prasugrel 1 50 -24 Bosutinib (SKI-606) 1 51 85 Nilotinib (AMN-107) 1 52 99 Enzastaurin (LY317615) 1 53 -12 Everolimus (RAD001) 1 54 94 Regorafenib (BAY 73-4506) 1 55 24 Thalidomide 1 56 40 Tivozanib (AV-951) 1 57 86 Fludarabine -

Sarcoma Metabolomics: Current Horizons and Future Perspectives
cells Review Sarcoma Metabolomics: Current Horizons and Future Perspectives Miguel Esperança-Martins 1,2,3,* , Isabel Fernandes 1,3,4 , Joaquim Soares do Brito 4,5, Daniela Macedo 6, Hugo Vasques 4,7, Teresa Serafim 2, Luís Costa 1,3,4 and Sérgio Dias 2,4 1 Centro Hospitalar Universitário Lisboa Norte, Medical Oncology Department, Hospital Santa Maria, 1649-028 Lisboa, Portugal; [email protected] (I.F.); [email protected] (L.C.) 2 Vascular Biology & Cancer Microenvironment Lab, Instituto de Medicina Molecular João Lobo Antunes, Faculdade de Medicina, Universidade de Lisboa, 1649-028 Lisboa, Portugal; tserafi[email protected] (T.S.); [email protected] (S.D.) 3 Translational Oncobiology Lab, Instituto de Medicina Molecular João Lobo Antunes, Faculdade de Medicina, Universidade de Lisboa, 1649-028 Lisboa, Portugal 4 Faculdade de Medicina, Universidade de Lisboa, 1649-028 Lisboa, Portugal; [email protected] (J.S.d.B.); [email protected] (H.V.) 5 Centro Hospitalar Universitário Lisboa Norte, Orthopedics and Traumatology Department, Hospital Santa Maria, 1649-028 Lisboa, Portugal 6 Medical Oncology Department, Hospital Lusíadas Lisboa, 1500-458 Lisboa, Portugal; [email protected] 7 General Surgery Department, Instituto Português de Oncologia de Lisboa Francisco Gentil, 1099-023 Lisboa, Portugal * Correspondence: [email protected] Abstract: The vast array of metabolic adaptations that cancer cells are capable of assuming, not Citation: Esperança-Martins, M.; only support their biosynthetic activity, but also fulfill their bioenergetic demands and keep their Fernandes, I.; Soares do Brito, J.; intracellular reduction–oxidation (redox) balance. Spotlight has recently been placed on the en- Macedo, D.; Vasques, H.; Serafim, T.; ergy metabolism reprogramming strategies employed by cancer cells to proliferate. -

Modifications to the Harmonized Tariff Schedule of the United States To
U.S. International Trade Commission COMMISSIONERS Shara L. Aranoff, Chairman Daniel R. Pearson, Vice Chairman Deanna Tanner Okun Charlotte R. Lane Irving A. Williamson Dean A. Pinkert Address all communications to Secretary to the Commission United States International Trade Commission Washington, DC 20436 U.S. International Trade Commission Washington, DC 20436 www.usitc.gov Modifications to the Harmonized Tariff Schedule of the United States to Implement the Dominican Republic- Central America-United States Free Trade Agreement With Respect to Costa Rica Publication 4038 December 2008 (This page is intentionally blank) Pursuant to the letter of request from the United States Trade Representative of December 18, 2008, set forth in the Appendix hereto, and pursuant to section 1207(a) of the Omnibus Trade and Competitiveness Act, the Commission is publishing the following modifications to the Harmonized Tariff Schedule of the United States (HTS) to implement the Dominican Republic- Central America-United States Free Trade Agreement, as approved in the Dominican Republic-Central America- United States Free Trade Agreement Implementation Act, with respect to Costa Rica. (This page is intentionally blank) Annex I Effective with respect to goods that are entered, or withdrawn from warehouse for consumption, on or after January 1, 2009, the Harmonized Tariff Schedule of the United States (HTS) is modified as provided herein, with bracketed matter included to assist in the understanding of proclaimed modifications. The following supersedes matter now in the HTS. (1). General note 4 is modified as follows: (a). by deleting from subdivision (a) the following country from the enumeration of independent beneficiary developing countries: Costa Rica (b). -

Targeting the Function of the HER2 Oncogene in Human Cancer Therapeutics
Oncogene (2007) 26, 6577–6592 & 2007 Nature Publishing Group All rights reserved 0950-9232/07 $30.00 www.nature.com/onc REVIEW Targeting the function of the HER2 oncogene in human cancer therapeutics MM Moasser Department of Medicine, Comprehensive Cancer Center, University of California, San Francisco, CA, USA The year 2007 marks exactly two decades since human HER3 (erbB3) and HER4 (erbB4). The importance of epidermal growth factor receptor-2 (HER2) was func- HER2 in cancer was realized in the early 1980s when a tionally implicated in the pathogenesis of human breast mutationally activated form of its rodent homolog neu cancer (Slamon et al., 1987). This finding established the was identified in a search for oncogenes in a carcinogen- HER2 oncogene hypothesis for the development of some induced rat tumorigenesis model(Shih et al., 1981). Its human cancers. An abundance of experimental evidence human homologue, HER2 was simultaneously cloned compiled over the past two decades now solidly supports and found to be amplified in a breast cancer cell line the HER2 oncogene hypothesis. A direct consequence (King et al., 1985). The relevance of HER2 to human of this hypothesis was the promise that inhibitors of cancer was established when it was discovered that oncogenic HER2 would be highly effective treatments for approximately 25–30% of breast cancers have amplifi- HER2-driven cancers. This treatment hypothesis has led cation and overexpression of HER2 and these cancers to the development and widespread use of anti-HER2 have worse biologic behavior and prognosis (Slamon antibodies (trastuzumab) in clinical management resulting et al., 1989). -
![Ehealth DSI [Ehdsi V2.2.2-OR] Ehealth DSI – Master Value Set](https://docslib.b-cdn.net/cover/8870/ehealth-dsi-ehdsi-v2-2-2-or-ehealth-dsi-master-value-set-1028870.webp)
Ehealth DSI [Ehdsi V2.2.2-OR] Ehealth DSI – Master Value Set
MTC eHealth DSI [eHDSI v2.2.2-OR] eHealth DSI – Master Value Set Catalogue Responsible : eHDSI Solution Provider PublishDate : Wed Nov 08 16:16:10 CET 2017 © eHealth DSI eHDSI Solution Provider v2.2.2-OR Wed Nov 08 16:16:10 CET 2017 Page 1 of 490 MTC Table of Contents epSOSActiveIngredient 4 epSOSAdministrativeGender 148 epSOSAdverseEventType 149 epSOSAllergenNoDrugs 150 epSOSBloodGroup 155 epSOSBloodPressure 156 epSOSCodeNoMedication 157 epSOSCodeProb 158 epSOSConfidentiality 159 epSOSCountry 160 epSOSDisplayLabel 167 epSOSDocumentCode 170 epSOSDoseForm 171 epSOSHealthcareProfessionalRoles 184 epSOSIllnessesandDisorders 186 epSOSLanguage 448 epSOSMedicalDevices 458 epSOSNullFavor 461 epSOSPackage 462 © eHealth DSI eHDSI Solution Provider v2.2.2-OR Wed Nov 08 16:16:10 CET 2017 Page 2 of 490 MTC epSOSPersonalRelationship 464 epSOSPregnancyInformation 466 epSOSProcedures 467 epSOSReactionAllergy 470 epSOSResolutionOutcome 472 epSOSRoleClass 473 epSOSRouteofAdministration 474 epSOSSections 477 epSOSSeverity 478 epSOSSocialHistory 479 epSOSStatusCode 480 epSOSSubstitutionCode 481 epSOSTelecomAddress 482 epSOSTimingEvent 483 epSOSUnits 484 epSOSUnknownInformation 487 epSOSVaccine 488 © eHealth DSI eHDSI Solution Provider v2.2.2-OR Wed Nov 08 16:16:10 CET 2017 Page 3 of 490 MTC epSOSActiveIngredient epSOSActiveIngredient Value Set ID 1.3.6.1.4.1.12559.11.10.1.3.1.42.24 TRANSLATIONS Code System ID Code System Version Concept Code Description (FSN) 2.16.840.1.113883.6.73 2017-01 A ALIMENTARY TRACT AND METABOLISM 2.16.840.1.113883.6.73 2017-01 -
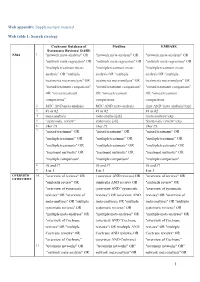
1 Web Appendix: Supplementary Material Web Table 1
Web appendix: Supplementary material Web table 1: Search strategy Cochrane Database of Medline EMBASE Systematic Reviews/ DARE NMA 1 "network meta-analysis" OR "network meta-analysis" OR "network meta-analysis" OR "network meta-regression" OR "network meta-regression" OR "network meta-regression" OR "multiple treatment meta- "multiple treatment meta- "multiple treatment meta- analysis” OR "multiple analysis OR "multiple analysis OR "multiple treatments meta-analysis" OR treatments meta-analysis" OR treatments meta-analysis" OR "mixed treatment comparison" "mixed treatment comparison" "mixed treatment comparison" OR "mixed treatment OR "mixed treatment OR "mixed treatment comparisons” comparisons comparisons 2 MTC AND meta-analysis MTC AND meta-analysis (mtc AND 'meta analysis'/exp) 3 #1 or #2 #1 or #2 #1 or #2 4 meta-analysis meta-analysis[sb] 'meta analysis'/exp 5 “systematic review” systematic [sb] 'Systematic review '/exp 6 #4or #5 #4or #5 #4or #5 7 "mixed treatment" OR "mixed treatment" OR "mixed treatment" OR "multiple treatment" OR "multiple treatment" OR "multiple treatment" OR "multiple treatments" OR "multiple treatments" OR "multiple treatments" OR "treatment networks" OR "treatment networks" OR "treatment networks" OR "multiple comparison" "multiple comparison" "multiple comparison" 8 #6 and #7 #6 and #7 #6 and #7 9 8 or 3 8 or 3 8 or 3 OVERVIEW 10 "overview of reviews" OR ( overview AND reviews) OR "overview of reviews" OR OF REVIEWS "umbrella review" OR (umbrella AND review) OR "umbrella review" OR "overview of systematic