BKM-React, an Integrated Biochemical Reaction Database Maren Lang†, Michael Stelzer† and Dietmar Schomburg*
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Next-Generation Sequencing of Representational Difference Analysis
Planta DOI 10.1007/s00425-017-2657-0 ORIGINAL ARTICLE Next-generation sequencing of representational difference analysis products for identification of genes involved in diosgenin biosynthesis in fenugreek (Trigonella foenum-graecum) 1 1 1 1 Joanna Ciura • Magdalena Szeliga • Michalina Grzesik • Mirosław Tyrka Received: 19 August 2016 / Accepted: 30 January 2017 Ó The Author(s) 2017. This article is published with open access at Springerlink.com Abstract Within the transcripts related to sterol and steroidal Main conclusion Representational difference analysis saponin biosynthesis, we discovered novel candidate of cDNA was performed and differential products were genes of diosgenin biosynthesis and validated their sequenced and annotated. Candidate genes involved in expression using quantitative RT-PCR analysis. Based on biosynthesis of diosgenin in fenugreek were identified. these findings, we supported the idea that diosgenin is Detailed mechanism of diosgenin synthesis was biosynthesized from cycloartenol via cholesterol. This is proposed. the first report on the next-generation sequencing of cDNA-RDA products. Analysis of the transcriptomes Fenugreek (Trigonella foenum-graecum L.) is a valuable enriched in low copy sequences contributed substantially medicinal and crop plant. It belongs to Fabaceae family to our understanding of the biochemical pathways of and has a unique potential to synthesize valuable steroidal steroid synthesis in fenugreek. saponins, e.g., diosgenin. Elicitation (methyl jasmonate) and precursor feeding (cholesterol and squalene) were Keywords Diosgenin Á Next-generation sequencing Á used to enhance the content of sterols and steroidal Phytosterols Á Representational difference analysis of sapogenins in in vitro grown plants for representational cDNA Á Steroidal saponins Á Transcriptome user-friendly difference analysis of cDNA (cDNA-RDA). -

Characterization of the Ergosterol Biosynthesis Pathway in Ceratocystidaceae
Journal of Fungi Article Characterization of the Ergosterol Biosynthesis Pathway in Ceratocystidaceae Mohammad Sayari 1,2,*, Magrieta A. van der Nest 1,3, Emma T. Steenkamp 1, Saleh Rahimlou 4 , Almuth Hammerbacher 1 and Brenda D. Wingfield 1 1 Department of Biochemistry, Genetics and Microbiology, Forestry and Agricultural Biotechnology Institute (FABI), University of Pretoria, Pretoria 0002, South Africa; [email protected] (M.A.v.d.N.); [email protected] (E.T.S.); [email protected] (A.H.); brenda.wingfi[email protected] (B.D.W.) 2 Department of Plant Science, University of Manitoba, 222 Agriculture Building, Winnipeg, MB R3T 2N2, Canada 3 Biotechnology Platform, Agricultural Research Council (ARC), Onderstepoort Campus, Pretoria 0110, South Africa 4 Department of Mycology and Microbiology, University of Tartu, 14A Ravila, 50411 Tartu, Estonia; [email protected] * Correspondence: [email protected]; Fax: +1-204-474-7528 Abstract: Terpenes represent the biggest group of natural compounds on earth. This large class of organic hydrocarbons is distributed among all cellular organisms, including fungi. The different classes of terpenes produced by fungi are mono, sesqui, di- and triterpenes, although triterpene ergosterol is the main sterol identified in cell membranes of these organisms. The availability of genomic data from members in the Ceratocystidaceae enabled the detection and characterization of the genes encoding the enzymes in the mevalonate and ergosterol biosynthetic pathways. Using Citation: Sayari, M.; van der Nest, a bioinformatics approach, fungal orthologs of sterol biosynthesis genes in nine different species M.A.; Steenkamp, E.T.; Rahimlou, S.; of the Ceratocystidaceae were identified. -

Catalase: Bioinformatics Analyses of One of the Key Enzymes in Hydrogen Peroxide Metabolism
6–71RYHPEHU 2019, Brno, Czech Republic Catalase: Bioinformatics analyses of one of the key enzymes in hydrogen peroxide metabolism Michaela Kameniarova, Romana Kopecka Department of Molecular Biology and Radiobiology Mendel University in Brno Zemedelska 1, 613 00 Brno CZECH REPUBLIC [email protected] Abstract: Catalases (CAT) are family of important antioxidant enzymes present in different isoforms and responsible for scavenging of hydrogen peroxide in almost all aerobically living organisms. In plants, they were found both in unicellular and multicellular species. Here, we performed bioinformatics analysis and analysed catalase evolutionary relationship and expression patterns. By comparing expression profiles of CATs and expression profiles of genes related to abiotic stimuli we found that almost 50% of light signalling genes were co-expressed with CATs. Further, by datamining in available resources and structural modelling we pinpointed candidate amino acid residues responsible for CAT thermostability. Key Words: catalase, H2O2, phylogenetics, abiotic stress, stability INTRODUCTION Plants contain several types of enzymes involved in H2O2 metabolism. These include catalases, ascorbate peroxidases, peroxiredoxins, glutathione/thioredoxin peroxidases, and glutathione S- transferases, but only catalases do not require additional cellular reductants (Mhamdi et al. 2010). Catalases are present almost in all aerobically respiring organisms. Within the cell environment, catalases are localized in all major sites of H2O2 production, mostly at peroxisomes, but they were detected in cytosol, mitochondria, and chloroplasts as well (Sharma and Ahmad 2014). Catalases (CAT, 1.11.1.6) are antioxidant enzymes that catalyse decomposition of hydrogen peroxide to water and molecular oxygen (2H2O2 → 2H2O + O2), an important process in defending cells against oxidative damage caused by reactive oxygen species (ROS; Alfonso-Prieto et al. -

Relating Metatranscriptomic Profiles to the Micropollutant
1 Relating Metatranscriptomic Profiles to the 2 Micropollutant Biotransformation Potential of 3 Complex Microbial Communities 4 5 Supporting Information 6 7 Stefan Achermann,1,2 Cresten B. Mansfeldt,1 Marcel Müller,1,3 David R. Johnson,1 Kathrin 8 Fenner*,1,2,4 9 1Eawag, Swiss Federal Institute of Aquatic Science and Technology, 8600 Dübendorf, 10 Switzerland. 2Institute of Biogeochemistry and Pollutant Dynamics, ETH Zürich, 8092 11 Zürich, Switzerland. 3Institute of Atmospheric and Climate Science, ETH Zürich, 8092 12 Zürich, Switzerland. 4Department of Chemistry, University of Zürich, 8057 Zürich, 13 Switzerland. 14 *Corresponding author (email: [email protected] ) 15 S.A and C.B.M contributed equally to this work. 16 17 18 19 20 21 This supporting information (SI) is organized in 4 sections (S1-S4) with a total of 10 pages and 22 comprises 7 figures (Figure S1-S7) and 4 tables (Table S1-S4). 23 24 25 S1 26 S1 Data normalization 27 28 29 30 Figure S1. Relative fractions of gene transcripts originating from eukaryotes and bacteria. 31 32 33 Table S1. Relative standard deviation (RSD) for commonly used reference genes across all 34 samples (n=12). EC number mean fraction bacteria (%) RSD (%) RSD bacteria (%) RSD eukaryotes (%) 2.7.7.6 (RNAP) 80 16 6 nda 5.99.1.2 (DNA topoisomerase) 90 11 9 nda 5.99.1.3 (DNA gyrase) 92 16 10 nda 1.2.1.12 (GAPDH) 37 39 6 32 35 and indicates not determined. 36 37 38 39 S2 40 S2 Nitrile hydration 41 42 43 44 Figure S2: Pearson correlation coefficients r for rate constants of bromoxynil and acetamiprid with 45 gene transcripts of ECs describing nucleophilic reactions of water with nitriles. -

( 12 ) United States Patent
US010208322B2 (12 ) United States Patent ( 10 ) Patent No. : US 10 ,208 , 322 B2 Coelho et al. (45 ) Date of Patent: * Feb . 19, 2019 ( 54 ) IN VIVO AND IN VITRO OLEFIN ( 56 ) References Cited CYCLOPROPANATION CATALYZED BY HEME ENZYMES U . S . PATENT DOCUMENTS 3 , 965 ,204 A 6 / 1976 Lukas et al. (71 ) Applicant: California Institute of Technology , 4 , 243 ,819 A 1 / 1981 Henrick Pasadena , CA (US ) 5 ,296 , 595 A 3 / 1994 Doyle 5 , 703 , 246 A 12 / 1997 Aggarwal et al. 7 , 226 , 768 B2 6 / 2007 Farinas et al. ( 72 ) Inventors : Pedro S . Coelho , Los Angeles, CA 7 , 267 , 949 B2 9 / 2007 Richards et al . (US ) ; Eric M . Brustad , Durham , NC 7 ,625 ,642 B2 12 / 2009 Matsutani et al. (US ) ; Frances H . Arnold , La Canada , 7 ,662 , 969 B2 2 / 2010 Doyle et al. CA (US ) ; Zhan Wang , San Jose , CA 7 ,863 ,030 B2 1 / 2011 Arnold (US ) ; Jared C . Lewis , Chicago , IL 8 ,247 ,430 B2 8 / 2012 Yuan 8 , 993 , 262 B2 * 3 / 2015 Coelho . .. .. .. • * • C12P 7 /62 (US ) 435 / 119 9 ,399 , 762 B26 / 2016 Farwell et al . (73 ) Assignee : California Institute of Technology , 9 , 493 ,799 B2 * 11 /2016 Coelho .. C12P 7162 Pasadena , CA (US ) 2006 / 0030718 AL 2 / 2006 Zhang et al. 2006 / 0111347 A1 5 / 2006 Askew , Jr . et al. 2007 /0276013 AL 11 /2007 Ebbinghaus et al . ( * ) Notice : Subject to any disclaimer , the term of this 2009 /0238790 A2 9 /2009 Sommadosi et al. patent is extended or adjusted under 35 2010 / 0056806 A1 3 / 2010 Warren U . -

BRENDA Tutorial
BRENDA Tutorial Introduction to the Enzyme Information System Facts about BRENDA (BRaunschweig ENzyme DAtabase) • one of the most comprehensive enzyme information repositories • BRENDA is a member of de.NBI (German Network for Bioinformatics Structure, since 2015) • BRENDA is an ELIXIR Core Data Resource (since 2018) • all enzymes, classified by the Enzyme Nomenclature (IUBMB) • data of molecular biology, biochemistry, medical research, and biotechnology • furthermore BRENDA includes data from interconnected databases containing results from text mining methods and bioinformatic approaches • BRENDA is freely available to the scientific community • more than 80,000 visits of the BRENDA website each month • major updates of the data in BRENDA are performed twice a year History and major developments of BRENDA • BRENDA was created at the former German National Research Center for Biotechnology (GBF, now HZI, Helmholtz Zentrum für Infektionsforschung, Braunschweig, Germany) in 1987 • BRENDA was originally published as a series of book o 1st Edition 1990-1997 (Enzyme Handbook) o 2nd Edition 2001-2013 (Handbook of Enzymes) • BRENDA moved to the University of Cologne, Germany • First online version in 1998 via the SRS system at the EBI • First website of BRENDA in Cologne • Transfer of BRENDA into a fully relational database system • BRENDA moved back to Braunschweig in 2007 • BRENDA is now maintained and further developed at the BRICS - TU Braunschweig Facts about BRENDA The main categories are based on the Enzymes and the Metabolites / Ligands Enzyme-related data encompasses information on: • Enzyme and ligand nomenclature • Organism • Reaction and specificity • Kinetic properties • Structure and role of the ligands • Stability information • Ligand-enzyme information • Enzyme sequence and structure • Mutants and disease • Occurrence, isolation, and preparation • Pathways BRENDA is the most comprehensive information system on: • 7862 EC Numbers (July 2019) • more than 2 Mill. -

Geneid T-Test Q-Val Gene Symbol Gene Title GO Biological Process
Supplementary Table 2. Genes selected by FADA as the most discriminant between tumoral and normal prostate samples GO Biological Process GO Molecular Function GO Cellular Component GeneID T-test q-val Gene Symbol Gene Title Description Description Description Pathway serine-type endopeptidase inhibitor activity /// endopeptidase inhibitor serpin peptidase inhibitor, clade activity /// serine-type endopeptidase 213572_s_at -6.753 2.213E-05 SERPINB1 B (ovalbumin), member 1 --- inhibitor activity cytoplasm --- ion transport /// cellular defense response /// positive regulation of T-cell, immune regulator 1, cell proliferation /// proton plasma membrane /// integral to ATPase, H+ transporting, transport /// transport /// proton transporter activity /// hydrogen ion plasma membrane /// membrane /// 204158_s_at -5.729 8.585E-05 TCIRG1 lysosomal V0 subunit A3 transport transporter activity integral to membrane --- chemotaxis /// cell adhesion /// homophilic cell adhesion /// nervous system development /// cell differentiation /// positive roundabout, axon guidance regulation of axonogenesis /// receptor activity /// axon guidance integral to plasma membrane /// cell receptor, homolog 1 development /// nervous system receptor activity /// identical protein surface /// membrane /// integral to 213194_at -6.168 4.441E-05 ROBO1 (Drosophila) development binding /// protein binding membrane --- structural molecule activity /// nucleus /// intermediate filament /// protein binding /// microtubule cytoplasmic dynein complex /// 203411_s_at -5.297 1.636E-04 -

Biological Role of Conceptus Derived Factors During Early Pregnancy In
Biological Role of Conceptus Derived Factors During Early Pregnancy in Ruminants A dissertation submitted in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY IN ANIMAL SCIENCES UNIVERSITY OF MISSOURI- COLUMBIA Division of Animal Science By KELSEY BROOKS Dr. Thomas Spencer, Dissertation Supervisor August 2016 The undersigned have examined the dissertation entitled, BIOLOGICAL ROLE OF CONCEPTUS DERIVED FACTORS DURING EARLY PREGNANCY IN RUMINANTS presented by Kelsey Brooks, a candidate for the degree of doctor of philosophy, and hereby certify that, in their opinion, it is worthy of acceptance. __________________________________ Chair, Dr. Thomas Spencer ___________________________________ Dr. Rodney Geisert ___________________________________ Dr. Randall Prather ___________________________________ Dr. Laura Schulz ACKNOWLEDGMENTS I would like to acknowledge all the students, faculty and staff at Washington State University and the University of Missouri for their help and support throughout my doctoral program. I am grateful for the opportunity to work with Dr. Thomas Spencer, and thank him for his input and guidance not only in planning experiments and completing projects but for helping me turn my love of science into a career in research. I would also like to acknowledge the members of my graduate committee at Washington State University for their help and input during the first 3 years of my studies. A special thanks to Dr. Jim Pru and Cindy Pru for providing unlimited entertainment, and the occasional missing reagent. Thank you to my committee members at the University of Missouri for adopting me late in my program and helping shape my future as an independent scientist. Thanks are also extended to members of the Prather lab and Wells lab for letting me in on the secrets of success using the CRISPR/Cas9 system. -

The Estimation of Kinetic Parameters in Systems Biology by Comparing Molecular Interaction Fields of Enzymes
237 Beilstein-Institut ESCEC, March 19th –23rd, 2006, Rdesheim/Rhein, Germany The Estimation of Kinetic Parameters in Systems Biology by Comparing Molecular Interaction Fields of Enzymes Matthias Stein, Razif R. Gabdoulline, Bruno Besson, Rebecca C. Wade EML Research gGmbH, Molecular and Cellular Modeling Group, Schloss-Wolfsbrunnenweg 33, 69118 Heidelberg, Germany E-Mail: [email protected] Received: 16th June 2006 / Published: 31st August 2007 Abstract The kinetic modelling of biochemical pathways requires a consistent set of enzymatic kinetic parameters. We report results from software development to assist the user in systems biology, allowing the retrie- val of heterogeneous protein sequence, structural and kinetic data. For the simulation of biological networks, missing enzymatic kinetic para- meters can be calculated using a similarity analysis of the enzymes’ molecular interaction fields. The quantitative PIPSA (qPIPSA) meth- odology relates changes in the molecular interaction fields of the enzymes with variations in the enzymatic rate constants or binding affinities. As an illustrative example, this approach is used to predict kinetic parameters for glucokinases from Escherichia coli based on experimental values for a test set of enzymes. The best correlation of the electrostatic potentials with kinetic parameters is found for the open form of the glucokinases. The similarity analysis was extended to a large set of glucokinases from various organisms. http://www.beilstein-institut.de/escec2006/proceedings/Stein/Stein.pdf 238 Stein, M. et al. Introduction One of the aims of systems biology is to provide a mathematical description of metabolic or signalling protein networks. This can be achieved by constructing a set of differential equations describing changes in concentrations of compounds with time [1]. -

Investigating the Role of ADP-Forming Acetyl-Coa Synthetase from The
Clemson University TigerPrints All Dissertations Dissertations 5-2016 Investigating the Role of ADP-forming Acetyl-CoA Synthetase from the Protozoan Parasite Entamoeba histolytica Cheryl Page Jones Clemson University, [email protected] Follow this and additional works at: https://tigerprints.clemson.edu/all_dissertations Recommended Citation Jones, Cheryl Page, "Investigating the Role of ADP-forming Acetyl-CoA Synthetase from the Protozoan Parasite Entamoeba histolytica" (2016). All Dissertations. 1668. https://tigerprints.clemson.edu/all_dissertations/1668 This Dissertation is brought to you for free and open access by the Dissertations at TigerPrints. It has been accepted for inclusion in All Dissertations by an authorized administrator of TigerPrints. For more information, please contact [email protected]. INVESTIGATING THE ROLE OF ADP-FORMING ACETYL-COA SYNTHETASE FROM THE PROTOZOAN PARASITE ENTAMOEBA HISTOLYTICA A Dissertation Presented to the Graduate School of Clemson University In Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy Biochemistry and Molecular Biology by Cheryl Page Jones May 2016 Accepted by Dr. Cheryl Ingram-Smith, Committee Chair Dr. William Marcotte, Jr. Dr. Julia Frugoli Dr. Kimberly Paul ABSTRACT ADP-forming acetyl-CoA synthetase (ACD; EC 6.2.1.13) catalyzes the reversible conversion of acetyl-CoA to acetate coupled to the production of ATP. This enzyme is present only in certain acetate-producing archaea and a limited number of bacteria and eukaryotes. ACD belongs to the same NDP-forming acyl-CoA synthetase enzyme superfamily as succinyl-CoA synthetase (SCS; EC 6.2.1.4) from the citric acid cycle, and a similar three-step mechanism involving a phosphoenzyme intermediate was originally proposed for this enzyme. -
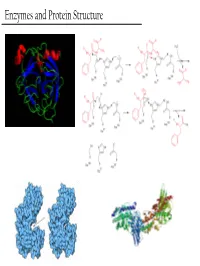
Enzymes and Protein Structure Last Week… PTM’S We (Re)Learned About
Enzymes and Protein Structure Last Week… PTM’s We (Re)Learned About Primary Structure And Tertiary Structure S-Q-D-A-G-M-Q-Q-G-A-D-M-D-Q-V-S-A Secondary Structure Enzymes What are these crazy things called ‘Enzymes’? And… Why do we care about them? - Enzymes are the catalysts of biology. TS They solve this problem This reaction will go, but it’ll take a G gazillion years… + OH- or You could heat it up to a gazillion degrees RC Enzymes Allow… - Higher reaction rates (same as any catalyst) Catalase - The rates of Enzyme reactions are 106 – 2H2O2 → 2H2O + O2 1012 times those of the corresponding Pretty darn fast: uncatalized reaction… ~ 6 * 105 sec-1 - The rates of reaction for the fastest enzymes are diffusion limited, around 106 turnovers per second. Enzymes Allow… - Milder reaction conditions (same as any catalyst, but better) - We won’t have to heat the reaction up to a gazillion degrees to make it go at a reasonable pace β-strand 4 β-strand 1 α-helix 2 β-strand 3 β-strand 2 - Enzymes from hythermophiles work well at around 85°C, but not at room temp!! β-strand 5 α-helix 1 Acylphosphatase from Sulfolobus solfataricus Enzymes Allow… - Reaction Specificity (better than inorganic catalysts) - Can’t really do this with an inorganic catalyst. A set of similar molecules will likely all be reactive… Enzymes Allow… - Capacity for Regulation (way better than inorganic catalysts) - For inorganic catalysts, you can really only interfere with their activity by using competitive inhibitors or permanent covalent deactivation E.g. -

SUPPLEMENTARY MATERIAL Supplementary Table 1
Predicting Drug Promiscuity Using Spherical Harmonic Surface The Open Conference Proceedings Journal, 2011, Volume 2 i SUPPLEMENTARY MATERIAL Supplementary Table 1. MDDR Target Annotations Used in the Promiscuity Predictions TARGET_KEY EXTERNAL_ID 1001 prostaglandin-endoperoxide synthase 1002 cyclooxygenase 1 1003 cyclooxygenase 2 1046 ribonucleoside-diphosphate reductase 1112 thymidylate synthase 1209 phosphoribosylglycinamide formyltransferase 1609 purine-nucleoside phosphorylase 1611 uridine phosphorylase 1637 nad adp-ribosyltransferase 1657 transferring alkyl or aryl groups, other than methyl groups 1798 thymidine kinase 1814 protein kinase 1843 1-phosphatidylinositol 4-kinase 1883 protein-tyrosine kinase 1973 nucleotidyltransferase 2018 rna-directed dna polymerase 2121 phospholipase a2 2124 acetylcholinesterase 2282 phosphoric diester hydrolase 2283 phosphodiesterase i 2309 3',5'-cyclic-nucleotide phosphodiesterase 2310 3',5'-cyclic-gmp phosphodiesterase 2316 steryl-sulfatase 2414 alpha-glucosidase 2565 adenosylhomocysteinase 2576 peptidase 25 aldehyde reductase 2613 dipeptidyl-peptidase iv 2619 peptidyl-dipeptidase a 2624 serine-type d-ala-d-ala carboxypeptidase 2659 serine endopeptidase 2663 trypsin 2664 thrombin 2665 coagulation factor xa 2672 coagulation factor viia 2695 tryptase 2742 cathepsin b 2750 cathepsin l 2765 caspase-1 2784 hiv-1 retropepsin 2811 metalloendopeptidase 2822 stromelysin 1 286 xanthine oxidase 2978 beta-lactamase 3014 adenosine deaminase ii The Open Conference Proceedings Journal, 2011, Volume 2 Pérez-Nueno