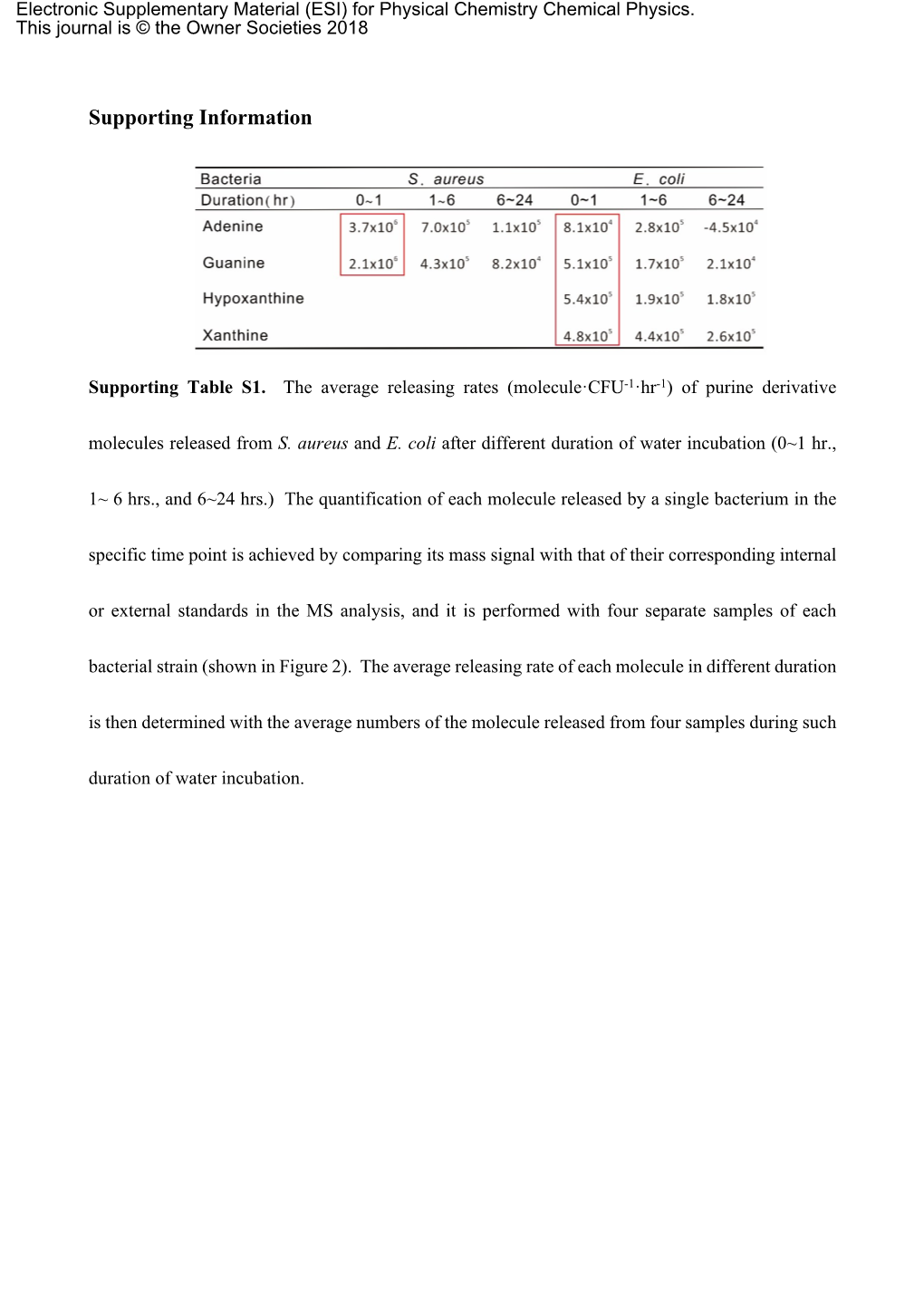
Supporting Information
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Alternative Biochemistries for Alien Life: Basic Concepts and Requirements for the Design of a Robust Biocontainment System in Genetic Isolation
G C A T T A C G G C A T genes Review Alternative Biochemistries for Alien Life: Basic Concepts and Requirements for the Design of a Robust Biocontainment System in Genetic Isolation Christian Diwo 1 and Nediljko Budisa 1,2,* 1 Institut für Chemie, Technische Universität Berlin Müller-Breslau-Straße 10, 10623 Berlin, Germany; [email protected] 2 Department of Chemistry, University of Manitoba, 144 Dysart Rd, 360 Parker Building, Winnipeg, MB R3T 2N2, Canada * Correspondence: [email protected] or [email protected]; Tel.: +49-30-314-28821 or +1-204-474-9178 Received: 27 November 2018; Accepted: 21 December 2018; Published: 28 December 2018 Abstract: The universal genetic code, which is the foundation of cellular organization for almost all organisms, has fostered the exchange of genetic information from very different paths of evolution. The result of this communication network of potentially beneficial traits can be observed as modern biodiversity. Today, the genetic modification techniques of synthetic biology allow for the design of specialized organisms and their employment as tools, creating an artificial biodiversity based on the same universal genetic code. As there is no natural barrier towards the proliferation of genetic information which confers an advantage for a certain species, the naturally evolved genetic pool could be irreversibly altered if modified genetic information is exchanged. We argue that an alien genetic code which is incompatible with nature is likely to assure the inhibition of all mechanisms of genetic information transfer in an open environment. The two conceivable routes to synthetic life are either de novo cellular design or the successive alienation of a complex biological organism through laboratory evolution. -

Geochemical Influences on Nonenzymatic Oligomerization Of
bioRxiv preprint doi: https://doi.org/10.1101/872234; this version posted December 11, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Geochemical influences on nonenzymatic oligomerization of prebiotically relevant cyclic nucleotides Authors: Shikha Dagar‡, Susovan Sarkar‡, Sudha Rajamani‡* ‡ Department of Biology, Indian Institute of Science Education and Research, Pune 411008, India Correspondence: [email protected]; Tel.: +91-20-2590-8061 Running title: Cyclic nucleotides and emergence of an RNA World Key words: Dehydration-rehydration cycles, lipid-assisted oligomerization, cyclic nucleotides, analogue environments Dagar, S. 1 bioRxiv preprint doi: https://doi.org/10.1101/872234; this version posted December 11, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract The spontaneous emergence of RNA on the early Earth continues to remain an enigma in the field of origins of life. Few studies have looked at the nonenzymatic oligomerization of cyclic nucleotides under neutral to alkaline conditions, in fully dehydrated state. Herein, we systematically investigated the oligomerization of cyclic nucleotides under prebiotically relevant conditions, where starting reactants were subjected to repeated dehydration-rehydration (DH- RH) regimes, like they would have been on an early Earth. DH-RH conditions, a recurring geological theme, are driven by naturally occurring processes including diurnal cycles and tidal pool activity. These conditions have been shown to facilitate uphill oligomerization reactions in terrestrial geothermal niches, which are hypothesized to be pertinent sites for the emergence of life. -

Nucleobases Thin Films Deposited on Nanostructured Transparent Conductive Electrodes for Optoelectronic Applications
www.nature.com/scientificreports OPEN Nucleobases thin flms deposited on nanostructured transparent conductive electrodes for optoelectronic applications C. Breazu1*, M. Socol1, N. Preda1, O. Rasoga1, A. Costas1, G. Socol2, G. Petre1,3 & A. Stanculescu1* Environmentally-friendly bio-organic materials have become the centre of recent developments in organic electronics, while a suitable interfacial modifcation is a prerequisite for future applications. In the context of researches on low cost and biodegradable resource for optoelectronics applications, the infuence of a 2D nanostructured transparent conductive electrode on the morphological, structural, optical and electrical properties of nucleobases (adenine, guanine, cytosine, thymine and uracil) thin flms obtained by thermal evaporation was analysed. The 2D array of nanostructures has been developed in a polymeric layer on glass substrate using a high throughput and low cost technique, UV-Nanoimprint Lithography. The indium tin oxide electrode was grown on both nanostructured and fat substrate and the properties of the heterostructures built on these two types of electrodes were analysed by comparison. We report that the organic-electrode interface modifcation by nano- patterning afects both the optical (transmission and emission) properties by multiple refections on the walls of nanostructures and the electrical properties by the efect on the organic/electrode contact area and charge carrier pathway through electrodes. These results encourage the potential application of the nucleobases thin flms deposited on nanostructured conductive electrode in green optoelectronic devices. Te use of natural or nature-inspired materials in organic electronics is a dynamic emerging research feld which aims to replace the synthesized materials with natural (bio) ones in organic electronics1–3. -

Nucleobase-Containing Compounds Evoke Behavioural, Olfactory, and Transcriptional Responses in Model Fishes
Correction-Nucleobase-containing compounds evoke behavioural, olfactory, and transcriptional responses in model fishes Shamchuk, A. L., Blunt, B. J., Lyons, D. D., Wang, M. Q., Gasheva, A., Lewis, C. R., Tomlin, K., Starr Hazard, E., Hardiman, G., & Tierney, K. B. (2018). Correction-Nucleobase-containing compounds evoke behavioural, olfactory, and transcriptional responses in model fishes. FACETS, 3(1). https://doi.org/10.1139/facets-2017-0101 Published in: FACETS Document Version: Publisher's PDF, also known as Version of record Queen's University Belfast - Research Portal: Link to publication record in Queen's University Belfast Research Portal Publisher rights Copyright 2018 the authors. This is an open access article published under a Creative Commons Attribution License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution and reproduction in any medium, provided the author and source are cited. 4.0), which permits unrestricted use, General rights Copyright for the publications made accessible via the Queen's University Belfast Research Portal is retained by the author(s) and / or other copyright owners and it is a condition of accessing these publications that users recognise and abide by the legal requirements associated with these rights. Take down policy The Research Portal is Queen's institutional repository that provides access to Queen's research output. Every effort has been made to ensure that content in the Research Portal does not infringe any person's rights, or applicable UK laws. If you discover content in the Research Portal that you believe breaches copyright or violates any law, please contact [email protected]. -

Development of a Universal Nucleobase and Modified
Development of a Universal Nucleobase and UNIT 1.5 Modified Nucleobases for Expanding the Genetic Code This unit presents protocols for the synthesis and characterization of nucleosides with unnatural bases in order to develop bases for the expansion of the genetic alphabet or for nonselective pairing opposite natural bases. The faithful pairing of nucleobases through complementary hydrogen-bond (H-bond) donors and acceptors forms the foundation of the genetic code. However, there is no reason to assume that the requirements for duplex stability and replication must limit the genetic alphabet to only two base pairs, or, for that matter, hydrogen-bonded base pairs. Expansion of this alphabet to contain a third base pair would allow for the encoding of additional information and would make possible a variety of in vitro experiments using nucleic acids with unnatural building blocks. Previous efforts to generate orthogonal base pairs have relied on H-bonding patterns that are not found with the canonical Watson-Crick pairs. However, in all cases, the unnatural bases were not kinetically orthogonal, and instead competitively paired with natural bases during polymerase-catalyzed DNA synthesis (Horlacher et al., 1995; Lutz et al., 1996, 1998a,b). Tautomeric isomerism, which would alter H-bond donor and acceptor patterns, likely contributes to this kinetic infidelity (Roberts et al., 1997a,b; Robinson et al., 1998; Beaussire and Pochet, 1999). An alternative strategy is centered around developing unnatural bases that form pairs based not on hydrogen bonds, but rather on interbase hydrophobic interactions. Such hydrophobic bases should not pair stably opposite natural bases due to the forced desolvation of the purines or pyrimidines. -

Overview of the Synthesis of Nucleoside Phosphates and Polyphosphates 13.1.6
Overview of the Synthesis of Nucleoside UNIT 13.1 Phosphates and Polyphosphates Phosphorylated nucleosides play a domi- ity to the synthesis. Side reactions can occur, nant role in biochemistry. Primary metabolism, such as depurination of the nucleoside, phos- DNA replication and repair, RNA synthesis, phorylation of the nucleobase, as well as chemi- protein synthesis, signal transduction, polysac- cal alteration of nucleobase analogs. Due to charide biosynthesis, and enzyme regulation their intrinsic reactivity, the synthesis of phos- are just a handful of processes involving these phoanhydride bonds is also synthetically chal- molecules. Literally thousands of enzymes use lenging. Phosphate anhydrides are phosphory- these compounds as substrates and/or regula- lating reagents that are readily degraded under tors. The need to obtain such compounds in acidic conditions. Finally, purification of syn- both labeled and unlabeled forms, as well as a thetic nucleotides can be problematic. Ionic burgeoning need for analogs, has driven the reagents, starting materials, and mixtures of development of a myriad of chemical and en- regioisomers (2′-, 3′-, 5′-phosphates) can be zymatic synthetic approaches. As chemical en- particularly difficult to separate from the de- tities, few molecules possess the wide array of sired product. densely packed functionality present in phos- In spite of the many potential difficulties phorylated nucleosides. This poses a formida- associated with nucleoside phosphorylation ble challenge to the synthetic chemist, one that and polyphosphorylation, a certain amount of has not yet been fully overcome. This overview success has been achieved in these areas. Given will address some common methods (synthetic the wealth of phosphorylating reagents avail- and enzymatic) used to construct phosphory- able, simple phosphorylation of nucleosides at lated nucleosides. -

Efforts and Challenges in Engineering the Genetic Code
life Review Efforts and Challenges in Engineering the Genetic Code Xiao Lin, Allen Chi Shing Yu and Ting Fung Chan * School of Life Sciences, The Chinese University of Hong Kong, Sha Tin, NT, Hong Kong, China; [email protected] (X.L.); [email protected] (A.C.S.Y.) * Correspondence: [email protected]; Tel.: +852-3943-6876 Academic Editor: Koji Tamura Received: 26 January 2017; Accepted: 10 March 2017; Published: 14 March 2017 Abstract: This year marks the 48th anniversary of Francis Crick’s seminal work on the origin of the genetic code, in which he first proposed the “frozen accident” hypothesis to describe evolutionary selection against changes to the genetic code that cause devastating global proteome modification. However, numerous efforts have demonstrated the viability of both natural and artificial genetic code variations. Recent advances in genetic engineering allow the creation of synthetic organisms that incorporate noncanonical, or even unnatural, amino acids into the proteome. Currently, successful genetic code engineering is mainly achieved by creating orthogonal aminoacyl-tRNA/synthetase pairs to repurpose stop and rare codons or to induce quadruplet codons. In this review, we summarize the current progress in genetic code engineering and discuss the challenges, current understanding, and future perspectives regarding genetic code modification. Keywords: frozen accident; genetic code; genetic engineering; evolution; synthetic biology 1. Introduction In 1968, Francis Crick first proposed the frozen accident theory of the genetic code [1]. The 20 canonical amino acids were once believed to be immutable elements of the code. The genetic code appears to be universal, from simple unicellular organisms to complex vertebrates. -

Nucleobase-Containing Polymers: Structure, Synthesis, and Applications
polymers Review Nucleobase-Containing Polymers: Structure, Synthesis, and Applications Haitao Yang 1 and Weixian Xi 2,3,* 1 School of Materials Science and Engineering, Nanchang Hangkong University, Nanchang 330063, China; [email protected] 2 Department of Chemical and Biomolecular Engineering, University of California Los Angeles, Los Angeles, CA 90095, USA 3 Department of Orthopedic Surgery, University of California Los Angeles, Los Angeles, CA 90095, USA * Correspondence: [email protected]; Tel.: +1-303-748-3805 Received: 10 November 2017; Accepted: 27 November 2017; Published: 1 December 2017 Abstract: Nucleobase interactions play a fundamental role in biological functions, including transcription and translation. Natural nucleic acids like DNA are also widely implemented in material realm such as DNA guided self-assembly of nanomaterials. Inspired by that, polymer chemists have contributed phenomenal endeavors to mimic both the structures and functions of natural nucleic acids in synthetic polymers. Similar sequence-dependent responses were observed and employed in the self-assembly of these nucleobase-containing polymers. Here, the structures, synthetic approaches, and applications of nucleobase-containing polymers are highlighted and a brief look is taken at the future development of these polymers. Keywords: nucleobase interactions; polymer synthesis; self-assembly 1. Introduction Nucleic acids in nature (DNA/RNA) represent the most powerful biomolecules. They serve as genetic material by hybridizing to a corresponding complementary strand to perform gene transcription and translation. Specifically, DNA carries genetic information from one generation to the next in living systems, and RNA guides the accurate assembly of amino acids into proteins by copying genetic information from DNA. Furthermore, in the materials science field, nucleic acids are also exploited for their selective hybridization capacities, unique geometric structures, ability to form nanoscale building blocks, directed self-assembly, and flexible programmability [1–4]. -

A DNA Pentaplex Incorporating Nucleobase Quintets
Proc. Natl. Acad. Sci. USA Vol. 96, pp. 10614–10619, September 1999 Biochemistry A DNA pentaplex incorporating nucleobase quintets JOHN C. CHAPUT AND CHRISTOPHER SWITZER† Department of Chemistry, University of California, Riverside, CA 92521 Edited by Leslie Orgel, The Salk Institute for Biological Studies, San Diego, CA, and approved July 7, 1999 (received for review April 29, 1999) ABSTRACT Supramolecular self-assembly is an integral ions. As a consequence, the iG-motif fulfills geometric pre- step in the formation of many biological structures. Here we dictions. report a DNA pentaplex that derives from a metal-assisted, hydrogen bond-mediated self-assembly process. In particular, cesium ions are found to induce pentameric assembly of DNA MATERIALS AND METHODS bearing the nonstandard nucleobase iso-guanine. The penta- plex was designed by using a simple algorithm to predict Oligonucleotides. Synthesis of iG phosphoramidite and oli- nucleobase structural requirements within a quintet motif. gonucleotides were performed as described (14). The design principles are general and should extend to PAGE Assays. Oligonucleotide (400 pmol) was combined complexes beyond pentaplex. Structures exhibiting molecu- with 1 lof10ϫ kinase buffer (700 mM Tris⅐HCl, pH ͞ ͞ ␥ 32 larities of five or more were previously accessible to peptides, 7.6 100 mM MgCl2 50 mM DTT), 1 lof - P-ATP (2 but not nucleic acids. Ci), 6 lofH2O, and 1 l of T4 polynucleotide kinase (3 units, United States Biochemical). The sample was incu- In abiologic systems, metal-mediated self-assembly has bated for 30 min at 37°C, at which time 1 lof660 MATP yielded arrays (1), nanometer-sized dendrimers (2), and was added, followed by incubation for an additional 30 min at 37°C. -

Nucleobase and Nucleoside Transport and Integration Into Plant Metabolism
REVIEW ARTICLE published: 09 September 2014 doi: 10.3389/fpls.2014.00443 Nucleobase and nucleoside transport and integration into plant metabolism Christopher Girke , Manuel Daumann , Sandra Niopek-Witz and Torsten Möhlmann* Pflanzenphysiologie, Fachbereich Biologie, Universität Kaiserslautern, Kaiserslautern, Germany Edited by: Nucleotide metabolism is an essential process in all living organisms. Besides newly Ilka Haferkamp, Universität synthesized nucleotides, the recycling (salvage) of partially degraded nucleotides, i.e., Kaiserslautern, Germany nucleosides and nucleobases serves to keep the homeostasis of the nucleotide pool. Reviewed by: Both types of metabolites are substrates of at least six families of transport proteins in Ian Kerr, University of Nottingham, UK Arabidopsis thaliana (Arabidopsis) with a total of 49 members. In the last years several Donato Pastore, University of members of such transport proteins have been analyzed allowing to present a more Foggia, Italy detailed picture of nucleoside and nucleobase transport and the physiological function of *Correspondence: these processes. Besides functioning in nucleotide metabolism it turned out that individual Torsten Möhlmann, members of the before named transporters exhibit the capacity to transport a wide range Pflanzenphysiologie, Universität Kaiserslautern, of different substrates including vitamins and phytohormones. The aim of this review is to Erwin-Schrödinger-Str., Postfach summarize the current knowledge on nucleobase and nucleoside transport processes in 3049, -

Alpha-Carboxy Nucleoside Phosphonates As Universal Nucleoside Triphosphate Mimics
Alpha-carboxy nucleoside phosphonates as universal nucleoside triphosphate mimics Jan Balzarinia,1, Kalyan Dasb,c, Jean A. Bernatchezd, Sergio E. Martinezb,c, Marianne Nguree,f, Sarah Keaneg, Alan Fordg, Nuala Maguireg, Niki Mullinsg, Jubi Johnh, Youngju Kimh, Wim Dehaenh, Johan Vande Voordea, Sandra Liekensa, Lieve Naesensa, Matthias Götted,e,f,i,2, Anita R. Maguireg, and Eddy Arnoldb,c aRega Institute for Medical Research and hDepartment of Chemistry, KU Leuven, B-3000 Leuven, Belgium; bCenter for Advanced Biotechnology and Medicine and cDepartment of Chemistry and Chemical Biology, Rutgers University, Piscataway, NJ 08854-8020; dDepartment of Biochemistry, McGill University, Montreal, QC H3G 1Y6, Canada; eDepartment of Microbiology and Immunology, McGill University, Montreal, QC H3A 2B4, Canada; fDepartment of Medical Microbiology and Immunology, University of Alberta, Edmonton, AB T6G 2E1, Canada; gDepartment of Chemistry and School of Pharmacy, Analytical and Biological Chemistry Research Facility, Synthesis and Solid State Pharmaceutical Centre, University College Cork, Cork, Ireland; and iDepartment of Medicine, Division of Experimental Medicine, McGill University, Montreal, QC H3A 1A3, Canada Edited by John M. Coffin, Tufts University School of Medicine, Boston, MA, and approved February 6, 2015 (received for review October 22, 2014) Polymerases have a structurally highly conserved negatively charged nucleobase part (i.e., to achieve optimal Watson–Crick base- + amino acid motif that is strictly required for Mg2 cation-depen- pairing with the template overhang), (ii) a replacement of the + dent catalytic incorporation of (d)NTP nucleotides into nucleic triphosphate moiety that should enable efficient Mg2 -directed acids. Based on these characteristics, a nucleoside monophospho- coordination, and (iii) a variable linker between the nucleobase nate scaffold, α-carboxy nucleoside phosphonate (α-CNP), was and the modified triphosphate to mimic the pentose entity present designed that is recognized by a variety of polymerases. -

A Model for the Emergence of RNA from a Prebiotically Plausible Mixture of Ribonucleotides, Arabinonucleotides and 2’-Deoxynucleotides
bioRxiv preprint doi: https://doi.org/10.1101/813675; this version posted October 22, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. A model for the emergence of RNA from a prebiotically plausible mixture of ribonucleotides, arabinonucleotides and 2’-deoxynucleotides. Seohyun Chris Kima,b, Lijun Zhoua,b, Wen Zhanga,b, Derek K. O’Flahertya,b,c, Valeria Rondo- Brovettoa,b, Jack W. Szostaka,b* aHoward Hughes Medical Institute, Department of Molecular Biology and Center for Computational and Integrative Biology, Massachusetts General Hospital, 185 Cambridge Street, Boston, Massachusetts 02114, United States. bDepartment of Genetics, Harvard Medical School, 77 Avenue Louis Pasteur, Boston, Massachusetts 02115, United States. cCurrent address: Alnylam Pharmaceuticals, 300 Third Street, Cambridge, Massachusetts 02142, USA *For correspondence: [email protected] bioRxiv preprint doi: https://doi.org/10.1101/813675; this version posted October 22, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract: The abiotic synthesis of ribonucleotides is thought to have been an essential step towards the emergence of the RNA world. However, it is likely that the prebiotic synthesis of ribonucleotides was accompanied by the simultaneous synthesis of arabinonucleotides, 2′-deoxyribonucleotides, and other variations on the canonical nucleotides. In order to understand how relatively homogeneous RNA could have emerged from such complex mixtures, we have examined the properties of arabinonucleotides and 2′- deoxyribonucleotides in nonenzymatic template-directed primer extension reactions.