Supporting Information
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Nitrate Prodrugs Able to Release Nitric Oxide in a Controlled and Selective
Europäisches Patentamt *EP001336602A1* (19) European Patent Office Office européen des brevets (11) EP 1 336 602 A1 (12) EUROPEAN PATENT APPLICATION (43) Date of publication: (51) Int Cl.7: C07C 205/00, A61K 31/00 20.08.2003 Bulletin 2003/34 (21) Application number: 02425075.5 (22) Date of filing: 13.02.2002 (84) Designated Contracting States: (71) Applicant: Scaramuzzino, Giovanni AT BE CH CY DE DK ES FI FR GB GR IE IT LI LU 20052 Monza (Milano) (IT) MC NL PT SE TR Designated Extension States: (72) Inventor: Scaramuzzino, Giovanni AL LT LV MK RO SI 20052 Monza (Milano) (IT) (54) Nitrate prodrugs able to release nitric oxide in a controlled and selective way and their use for prevention and treatment of inflammatory, ischemic and proliferative diseases (57) New pharmaceutical compounds of general effects and for this reason they are useful for the prep- formula (I): F-(X)q where q is an integer from 1 to 5, pref- aration of medicines for prevention and treatment of in- erably 1; -F is chosen among drugs described in the text, flammatory, ischemic, degenerative and proliferative -X is chosen among 4 groups -M, -T, -V and -Y as de- diseases of musculoskeletal, tegumental, respiratory, scribed in the text. gastrointestinal, genito-urinary and central nervous sys- The compounds of general formula (I) are nitrate tems. prodrugs which can release nitric oxide in vivo in a con- trolled and selective way and without hypotensive side EP 1 336 602 A1 Printed by Jouve, 75001 PARIS (FR) EP 1 336 602 A1 Description [0001] The present invention relates to new nitrate prodrugs which can release nitric oxide in vivo in a controlled and selective way and without the side effects typical of nitrate vasodilators drugs. -
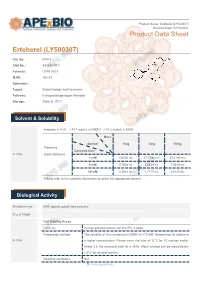
Erteberel (LY500307) Product Data Sheet
Product Name: Erteberel (LY500307) Revision Date: 01/10/2021 Product Data Sheet Erteberel (LY500307) Cat. No.: B1518 CAS No.: 533884-09-2 Formula: C18H18O3 M.Wt: 282.33 Synonyms: Target: Endocrinology and Hormones Pathway: Estrogen/progestogen Receptor Storage: Store at -20°C Solvent & Solubility insoluble in H2O; ≥14.1 mg/mL in DMSO; ≥48.3 mg/mL in EtOH Mass Solvent 1mg 5mg 10mg Preparing Concentration In Vitro Stock Solutions 1 mM 3.5420 mL 17.7098 mL 35.4195 mL 5 mM 0.7084 mL 3.5420 mL 7.0839 mL 10 mM 0.3542 mL 1.7710 mL 3.5420 mL Please refer to the solubility information to select the appropriate solvent. Biological Activity Shortsummary ERβ agonist, potent and selective IC₅₀ & Target Cell Viability Assay Cell Line: Human prostate cancer cell line (PC-3 cells) Preparation method: The solubility of this compound in DMSO is >10 mM. General tips for obtaining In Vitro a higher concentration: Please warm the tube at 37°C for 10 minutes and/or shake it in the ultrasonic bath for a while. Stock solution can be stored below -20°C for several months. Reacting conditions: N/A 1 | www.apexbt.com Applications: Erteberel showed potent and selective binding affinity for ERβ with EC50 value of 0.66 nM [1]. Animal experiment Animal models: Male and female rat fertility and rat and rabbit embryo-fetal development model Dosage form: 0.03 to 10 mg/kg/day for rats, or 1 to 25 mg/kg/day for rabbits, oral gavage, for 2 or 10 weeks Applications: There were no-observed adverse effect levels following LY500307 In Vivo administration of 1 mg/kg/day for male rat fertility, 0.3 mg/kg/day for female rat fertility and embryo-fetal development, and 25 mg/kg/day for rabbit embryo-fetal development [2]. -

Supplementary Information
Supplementary Information Network-based Drug Repurposing for Novel Coronavirus 2019-nCoV Yadi Zhou1,#, Yuan Hou1,#, Jiayu Shen1, Yin Huang1, William Martin1, Feixiong Cheng1-3,* 1Genomic Medicine Institute, Lerner Research Institute, Cleveland Clinic, Cleveland, OH 44195, USA 2Department of Molecular Medicine, Cleveland Clinic Lerner College of Medicine, Case Western Reserve University, Cleveland, OH 44195, USA 3Case Comprehensive Cancer Center, Case Western Reserve University School of Medicine, Cleveland, OH 44106, USA #Equal contribution *Correspondence to: Feixiong Cheng, PhD Lerner Research Institute Cleveland Clinic Tel: +1-216-444-7654; Fax: +1-216-636-0009 Email: [email protected] Supplementary Table S1. Genome information of 15 coronaviruses used for phylogenetic analyses. Supplementary Table S2. Protein sequence identities across 5 protein regions in 15 coronaviruses. Supplementary Table S3. HCoV-associated host proteins with references. Supplementary Table S4. Repurposable drugs predicted by network-based approaches. Supplementary Table S5. Network proximity results for 2,938 drugs against pan-human coronavirus (CoV) and individual CoVs. Supplementary Table S6. Network-predicted drug combinations for all the drug pairs from the top 16 high-confidence repurposable drugs. 1 Supplementary Table S1. Genome information of 15 coronaviruses used for phylogenetic analyses. GenBank ID Coronavirus Identity % Host Location discovered MN908947 2019-nCoV[Wuhan-Hu-1] 100 Human China MN938384 2019-nCoV[HKU-SZ-002a] 99.99 Human China MN975262 -

The Role of Sevista in the Management of Dysfunctional Uterine Bleeding Section Obstetrics & Gynaecology
Original Article DOI: 10.7860/JCDR/2012/4794.2687 The Role of Sevista in the Management of Dysfunctional Uterine Bleeding Section Obstetrics & Gynaecology DHANANJAY BS, SUNIL KUMAR NANDA ABSTRACT bleeding, a diagnosis of DUB was made and the treatment with Objective: The complaints of excessive menstrual bleeding ormiloxifene was started. The number of cases were 35 cases. (menorrhagia) have a substantial impact on the gynaecological The treatment with ormeloxifene was evaluated by measuring services and in most of the cases, no organic pathology is iden- the Hb g/dl and the endometrial thickness before and after 3 tified. Nonsteroidal anti-inflammatory drugs and tranexamic acid months of treatment with sevista. Ormeloxifene was given in the offer a simple therapy which has to be taken during menses, with dosage of a 60 mg tablet twice a week for 3 months, followed by reductions of 25-35% and 50% respectively in the Menstrual once a week for another 3 months. Blood Loss (MBL). Danazol and the gonadatrophin-releasing Observation & Results: There was a statistically significant in- hormone analogues are highly effective, but their side-effects crease in the Hb g/dl (p < 0.001) and a statistically significant make them suitable only for a short-term use. In the present decrease in the endometrial thickness (p< 0.001) after the treat- study, the role of ormeloxifene was studied in patients of DUB. ment with ormeloxifene. Materials & Methods: The subjects were diagnosed cases of Conclusion: Ormeloxifene can be used asa effective drug in the DUB. After ruling out the possible causes of the abnormal uterine treatment of Dysfunctional uterine bleeding. -

Tanibirumab (CUI C3490677) Add to Cart
5/17/2018 NCI Metathesaurus Contains Exact Match Begins With Name Code Property Relationship Source ALL Advanced Search NCIm Version: 201706 Version 2.8 (using LexEVS 6.5) Home | NCIt Hierarchy | Sources | Help Suggest changes to this concept Tanibirumab (CUI C3490677) Add to Cart Table of Contents Terms & Properties Synonym Details Relationships By Source Terms & Properties Concept Unique Identifier (CUI): C3490677 NCI Thesaurus Code: C102877 (see NCI Thesaurus info) Semantic Type: Immunologic Factor Semantic Type: Amino Acid, Peptide, or Protein Semantic Type: Pharmacologic Substance NCIt Definition: A fully human monoclonal antibody targeting the vascular endothelial growth factor receptor 2 (VEGFR2), with potential antiangiogenic activity. Upon administration, tanibirumab specifically binds to VEGFR2, thereby preventing the binding of its ligand VEGF. This may result in the inhibition of tumor angiogenesis and a decrease in tumor nutrient supply. VEGFR2 is a pro-angiogenic growth factor receptor tyrosine kinase expressed by endothelial cells, while VEGF is overexpressed in many tumors and is correlated to tumor progression. PDQ Definition: A fully human monoclonal antibody targeting the vascular endothelial growth factor receptor 2 (VEGFR2), with potential antiangiogenic activity. Upon administration, tanibirumab specifically binds to VEGFR2, thereby preventing the binding of its ligand VEGF. This may result in the inhibition of tumor angiogenesis and a decrease in tumor nutrient supply. VEGFR2 is a pro-angiogenic growth factor receptor -

WHO Drug Information Vol
WHO Drug Information Vol. 24, No. 4, 2010 World Health Organization WHO Drug Information Contents WHO Prequalification Sitaxentan: worldwide withdrawal 307 Programmes Sibutramine: suspension of sales 307 Sibutramine-containing medicines: WHO Prequalification of Medicines withdrawal 308 Programme: survey of service Testosterone transdermal patch: quality provided to manufacturers 293 withdrawal of extension of WHO initiates pilot prequalification of indication application 308 active pharmaceutical ingredients 297 Aliskiren/valsartan: withdrawal of New on-line database for WHO marketing authorization application 308 prequalified vaccines 298 Mometasone furoate/formoterol fumarate: withdrawal of marketing Safety and Efficacy Issues authorization application 309 EMA and US FDA extend confidentiality H1N1 influenza vaccine: narcolepsy 299 arrangements indefinitely 309 Statins: interstitial lung disease 299 Tocilizumab: risk of fatal anaphylaxis 300 Recent Publications, Pioglitazone: potential bladder cancer 301 Information and Events Angiotensin receptor blockers and US Government to share patents with cancer: safety review 301 Medicines Patent Pool 310 GnRH agonists, diabetes and cardio- Clinical trials and global medicines vascular disease 301 development 310 Gadolinium-based contrast agents: Evaluation of future nanomedicines 311 kidney dysfunction 302 Reporting on opioid inaccessibility 311 Lamotrigine: aseptic meningitis Tinzaparin sodium: renal Impairment in elderly 303 Consultation Documents Tamoxifen: drug interactions involving The -

TE INI (19 ) United States (12 ) Patent Application Publication ( 10) Pub
US 20200187851A1TE INI (19 ) United States (12 ) Patent Application Publication ( 10) Pub . No .: US 2020/0187851 A1 Offenbacher et al. (43 ) Pub . Date : Jun . 18 , 2020 ( 54 ) PERIODONTAL DISEASE STRATIFICATION (52 ) U.S. CI. AND USES THEREOF CPC A61B 5/4552 (2013.01 ) ; G16H 20/10 ( 71) Applicant: The University of North Carolina at ( 2018.01) ; A61B 5/7275 ( 2013.01) ; A61B Chapel Hill , Chapel Hill , NC (US ) 5/7264 ( 2013.01 ) ( 72 ) Inventors: Steven Offenbacher, Chapel Hill , NC (US ) ; Thiago Morelli , Durham , NC ( 57 ) ABSTRACT (US ) ; Kevin Lee Moss, Graham , NC ( US ) ; James Douglas Beck , Chapel Described herein are methods of classifying periodontal Hill , NC (US ) patients and individual teeth . For example , disclosed is a method of diagnosing periodontal disease and / or risk of ( 21) Appl. No .: 16 /713,874 tooth loss in a subject that involves classifying teeth into one of 7 classes of periodontal disease. The method can include ( 22 ) Filed : Dec. 13 , 2019 the step of performing a dental examination on a patient and Related U.S. Application Data determining a periodontal profile class ( PPC ) . The method can further include the step of determining for each tooth a ( 60 ) Provisional application No.62 / 780,675 , filed on Dec. Tooth Profile Class ( TPC ) . The PPC and TPC can be used 17 , 2018 together to generate a composite risk score for an individual, which is referred to herein as the Index of Periodontal Risk Publication Classification ( IPR ) . In some embodiments , each stage of the disclosed (51 ) Int. Cl. PPC system is characterized by unique single nucleotide A61B 5/00 ( 2006.01 ) polymorphisms (SNPs ) associated with unique pathways , G16H 20/10 ( 2006.01 ) identifying unique druggable targets for each stage . -

Modifications to the Harmonized Tariff Schedule of the United States To
U.S. International Trade Commission COMMISSIONERS Shara L. Aranoff, Chairman Daniel R. Pearson, Vice Chairman Deanna Tanner Okun Charlotte R. Lane Irving A. Williamson Dean A. Pinkert Address all communications to Secretary to the Commission United States International Trade Commission Washington, DC 20436 U.S. International Trade Commission Washington, DC 20436 www.usitc.gov Modifications to the Harmonized Tariff Schedule of the United States to Implement the Dominican Republic- Central America-United States Free Trade Agreement With Respect to Costa Rica Publication 4038 December 2008 (This page is intentionally blank) Pursuant to the letter of request from the United States Trade Representative of December 18, 2008, set forth in the Appendix hereto, and pursuant to section 1207(a) of the Omnibus Trade and Competitiveness Act, the Commission is publishing the following modifications to the Harmonized Tariff Schedule of the United States (HTS) to implement the Dominican Republic- Central America-United States Free Trade Agreement, as approved in the Dominican Republic-Central America- United States Free Trade Agreement Implementation Act, with respect to Costa Rica. (This page is intentionally blank) Annex I Effective with respect to goods that are entered, or withdrawn from warehouse for consumption, on or after January 1, 2009, the Harmonized Tariff Schedule of the United States (HTS) is modified as provided herein, with bracketed matter included to assist in the understanding of proclaimed modifications. The following supersedes matter now in the HTS. (1). General note 4 is modified as follows: (a). by deleting from subdivision (a) the following country from the enumeration of independent beneficiary developing countries: Costa Rica (b). -

Screening of Clinically Approved and Investigation Drugs As Potential
Screening of Clinically Approved and Investigation Drugs as Potential Inhibitors of COVID-19 Main Protease: A Virtual Drug Repurposing Study Serdar Durdagi1,*, Busecan Aksoydan1,2, Berna Dogan1, Kader Sahin1, Aida Shahraki1,3 1Computational Biology and Molecular Simulations Laboratory, Department of Biophysics, School of Medicine, Bahcesehir University, Istanbul, Turkey 2Neuroscience Program, Graduate School of Health Sciences, Bahçeşehir University, Istanbul, Turkey 3Department of Molecular Biology and Genetics, Bogazici University, Istanbul, Turkey Abstract There is an urgent need for a new drug against COVID-19. Since designing a new drug and testing its pharmacokinetics and pharmacodynamics properties may take years, here we used a physics-driven high throughput virtual screening drug re-purposing approach to identify new compounds against COVID-19. As the molecules considered in repurposing studies passed through several stages and have well-defined profiles, they would not require prolonged pre- clinical studies and hence, they would be excellent candidates in the cases of disease emergencies or outbreaks. While the spike protein is the key for the virus to enter the cell though the interaction with ACE2, enzymes such as main protease are crucial for the life cycle of the virus. This protein is one of the most attractive targets for the development of new drugs against COVID-19 due to its pivotal role in the replication and transcription of the virus. We used 7922 FDA approved small molecule drugs as well as compounds in clinical investigation from NIH Chemical Genomics Center (NCGC) Pharmaceutical Collection (NPC) database in our drug repurposing study. Both apo and holo forms of target protein COVID-19 main proteases were used in virtual screening. -

Mitochondrial Estrogen Receptors Alter Mitochondrial Priming and Response to Endocrine Therapy in Breast Cancer Cells
Mitochondrial estrogen receptors alter mitochondrial priming and response to endocrine therapy in breast cancer cells. Ozgur Kutuk ( [email protected] ) Başkent University, Adana Dr. Turgut Noyan Medical and Research Center https://orcid.org/0000-0001- 9854-7220 Bahriye Karakas Sabanci University Yeliz Aka Baskent University Asli Giray Alanya Alaaddin Keykubat University Huveyda Basaga Sabanci University Ufuk Acikbas Baskent University Ozgur Gul Bilgi University Sehime Gulsun Temel Uludag University Article Keywords: breast cancer, BH3, Estrogen receptor, mitochondrial estrogen signaling Posted Date: February 5th, 2021 DOI: https://doi.org/10.21203/rs.3.rs-139821/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 1/29 Abstract Breast cancer is the most common cancer with a high rate of mortality and morbidity among women worldwide. Estrogen receptor status is an important prognostic factor and endocrine therapy is the choice of rst-line treatment in ER-positive breast cancer. However, most tumors develop resistance to endocrine therapy. Here we demonstrate that BH3 proling technology, in particular dynamic BH3 proling can predict the response to endocrine therapy agents as well as development of acquired resistance in breast cancer cells independent of estrogen receptor status. Immunouorescence analysis and subcellular fractionation experiments revealed distinct ER-α and ER-β subcellular localization patterns in breast cancer cells, including mitochondrial localization of both receptor subtypes. shRNA-mediated depletion of ER-β in breast cancer cells led to resistance to endocrine therapy agents and selective reconstitution of ER-β in mitochondria restored sensitivity. Notably, mitochondria-targeted ER-α did not restore sensitivity, even conferred further resistance to endocrine therapy agents. -

Mitochondrial Estrogen Receptors Alter Mitochondrial Priming and Response to Endocrine Therapy in Breast Cancer Cells
www.nature.com/cddiscovery ARTICLE OPEN Mitochondrial estrogen receptors alter mitochondrial priming and response to endocrine therapy in breast cancer cells ✉ Bahriye Karakas1, Yeliz Aka2, Asli Giray3, Sehime Gulsun Temel4,5,6, Ufuk Acikbas2, Huveyda Basaga1, Ozgur Gul7 and Ozgur Kutuk 2 © The Author(s) 2021 Breast cancer is the most common cancer with a high rate of mortality and morbidity among women worldwide. Estrogen receptor status is an important prognostic factor and endocrine therapy is the choice of first-line treatment in ER-positive breast cancer. However, most tumors develop resistance to endocrine therapy. Here we demonstrate that BH3 profiling technology, in particular, dynamic BH3 profiling can predict the response to endocrine therapy agents as well as the development of acquired resistance in breast cancer cells independent of estrogen receptor status. Immunofluorescence analysis and subcellular fractionation experiments revealed distinct ER-α and ER-β subcellular localization patterns in breast cancer cells, including mitochondrial localization of both receptor subtypes. shRNA-mediated depletion of ER-β in breast cancer cells led to resistance to endocrine therapy agents and selective reconstitution of ER-β in mitochondria restored sensitivity. Notably, mitochondria-targeted ER-α did not restore sensitivity, even conferred further resistance to endocrine therapy agents. In addition, expressing mitochondria-targeted ER-β in breast cancer cells resulted in decreased mitochondrial respiration alongside increased total ROS and mitochondrial superoxide production. Furthermore, our data demonstrated that mitochondrial ER-β can be successfully targeted by the selective ER-β agonist Erteberel. Thus, our findings provide novel findings on mitochondrial estrogen signaling in breast cancer cells and suggest the implementation of the dynamic BH3 technique as a tool to predict acquired endocrine therapy resistance. -

In Silico Prediction of Heliannuol A, B, C, D, and E Compounds on Estrogen Receptor Β Agonists
Indonesian Journal of Cancer Chemoprevention, February 2021 ISSN: 2088–0197 e-ISSN: 2355-8989 In Silico Prediction of Heliannuol A, B, C, D, and E Compounds on Estrogen Receptor β Agonists Roihatul Mutiah, Alif Firman Firdausy, Yen Yen Ari Indrawijaya, Hibbatullah* Department of Pharmacy, Faculty of Medical and Health Sciences, Maulana Malik Ibrahim State Islamic University of Malang, Indonesia Abstract Heliannuols has a benzoxepine ring that produces anticancer activity by the inhibition mechanism of phosphoinositide 3 kinases (PI3K). Heliannuols are a compound that can be found in the leaves of sunflower Helianthus( annuus L.). The purpose of this study is to predict interactions, toxicity, physicochemical, and pharmacokinetics of Heliannuol A, B, C, D, and E based in silico as candidate anticancer drugs. Estrogen receptor beta (ERβ) is a new potential therapy for glioma with an antiproliferative effect. Ligands agonist ERβ have the potential activity to inhibit the proliferation of glioma cells and the discovery of this ligand has opened new therapy through the ERβ to prolong survival in cancer patients. Prediction of physicochemical properties based on Lipinski rules and penetrate in the blood-brain barrier. Receptor validation shows that 2I0G(A) has a smaller RMSD value than 2I0G(B), receptor validation is valid if the RMSD value less than 2. The result of molecular docking shows that Heliannuols comply with Lipinski rules and have low toxicity. Heliannuols also have a similar amino acid with comparison drug (Erteberel), but the rerank score of Erteberel still lower than Heliannuols. Keywords: Helianthus annuus, Heliannuols, estrogen receptor β (ERβ), in silico, toxicity. INTRODUCTION using HPLC with hexane and ethyl acetate solvents (Macias, et al., 1994; Macias, et al., 2000; Macias, Sunflower plant (Helianthus annuus L.) has et al., 2002).