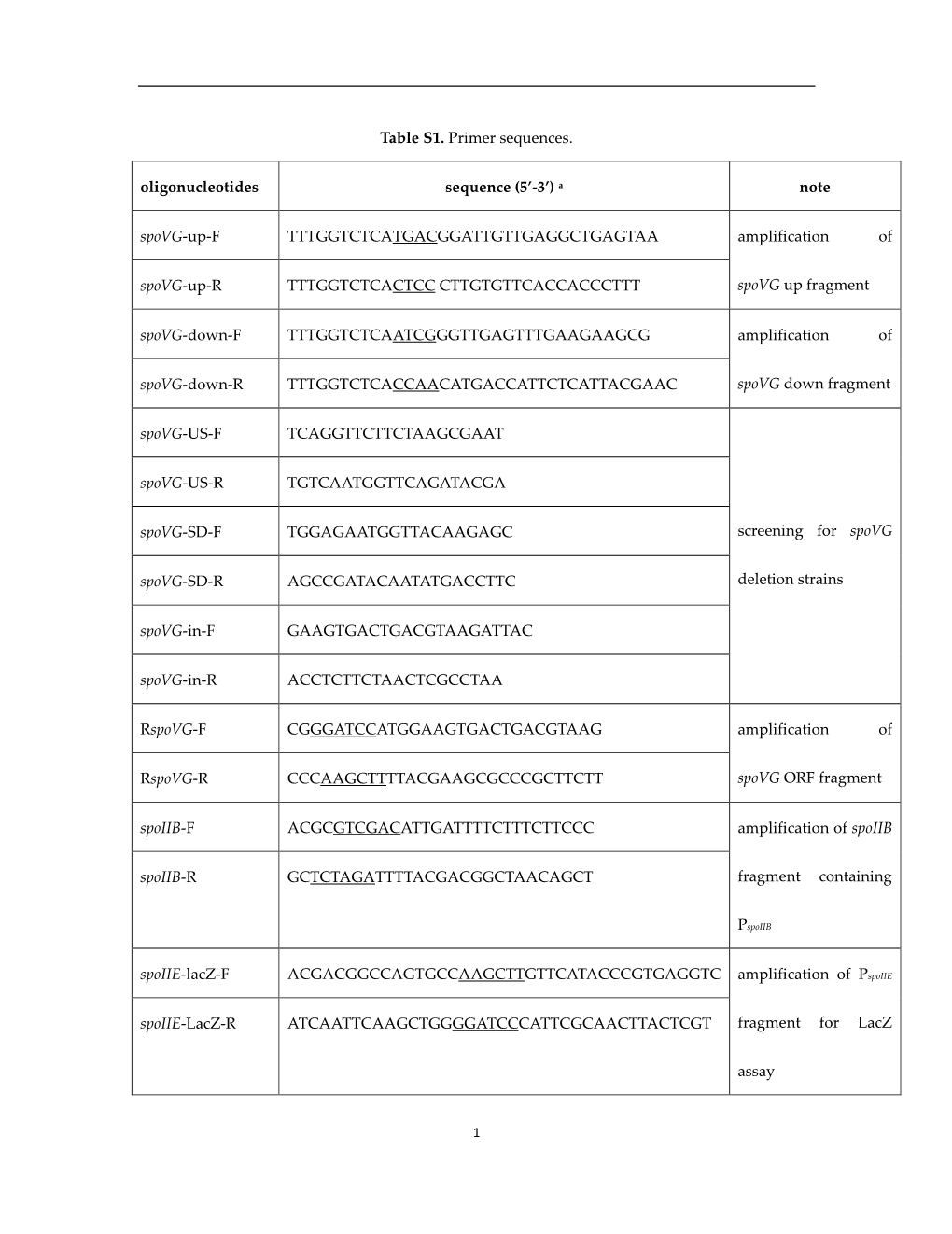
Table S1. Primer Sequences. Oligonucleotides Sequence (5'-3') A
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

METACYC ID Description A0AR23 GO:0004842 (Ubiquitin-Protein Ligase
Electronic Supplementary Material (ESI) for Integrative Biology This journal is © The Royal Society of Chemistry 2012 Heat Stress Responsive Zostera marina Genes, Southern Population (α=0. -

Characterization of L-Serine Deaminases, Sdaa (PA2448) and Sdab 2 (PA5379), and Their Potential Role in Pseudomonas Aeruginosa 3 Pathogenesis 4 5 Sixto M
bioRxiv preprint doi: https://doi.org/10.1101/394957; this version posted August 20, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 Characterization of L-serine deaminases, SdaA (PA2448) and SdaB 2 (PA5379), and their potential role in Pseudomonas aeruginosa 3 pathogenesis 4 5 Sixto M. Leal1,6, Elaine Newman2 and Kalai Mathee1,3,4,5 * 6 7 Author affiliations: 8 9 1Department of Biological Sciences, College of Arts Sciences and 10 Education, Florida International University, Miami, United States of 11 America 12 2Department of Biological Sciences, Concordia University, Montreal, 13 Canada 14 3Department of Molecular Microbiology and Infectious Diseases, Herbert 15 Wertheim College of Medicine, Florida International University, Miami, 16 United States of America 17 4Biomolecular Sciences Institute, Florida International University, Miami, 18 United States of America 19 20 Present address: 21 22 5Department of Human and Molecular Genetics, Herbert Wertheim 23 College of Medicine, Florida International University, Miami, United States 24 of America 25 6Case Western Reserve University, United States of America 26 27 28 *Correspondance: Kalai Mathee, MS, PhD, 29 [email protected] 30 31 Telephone : 1-305-348-0628 32 33 Keywords: Serine Catabolism, Central Metabolism, TCA Cycle, Pyruvate, 34 Leucine Responsive Regulatory Protein (LRP), One Carbon Metabolism 35 Running title: P. aeruginosa L-serine deaminases 36 Subject category: Pathogenicity and Virulence/Host Response 37 1 bioRxiv preprint doi: https://doi.org/10.1101/394957; this version posted August 20, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. -

QM/MM) Study on Ornithine Cyclodeaminase (OCD): a Tale of Two Iminiums
Int. J. Mol. Sci. 2012, 13, 12994-13011; doi:10.3390/ijms131012994 OPEN ACCESS International Journal of Molecular Sciences ISSN 1422-0067 www.mdpi.com/journal/ijms Article A Molecular Dynamics (MD) and Quantum Mechanics/Molecular Mechanics (QM/MM) Study on Ornithine Cyclodeaminase (OCD): A Tale of Two Iminiums Bogdan F. Ion, Eric A. C. Bushnell, Phil De Luna and James W. Gauld * Department of Chemistry and Biochemistry, University of Windsor, Windsor, ON N9B 3P4, Canada; E-Mails: [email protected] (B.F.I.); [email protected] (E.A.C.B.); [email protected] (P.D.L.) * Author to whom correspondence should be addressed; E-Mail: [email protected]; Tel.: +1-519-253-3000; Fax: +1-519-973-7098. Received: 10 September 2012; in revised form: 27 September 2012 / Accepted: 27 September 2012 / Published: 11 October 2012 Abstract: Ornithine cyclodeaminase (OCD) is an NAD+-dependent deaminase that is found in bacterial species such as Pseudomonas putida. Importantly, it catalyzes the direct conversion of the amino acid L-ornithine to L-proline. Using molecular dynamics (MD) and a hybrid quantum mechanics/molecular mechanics (QM/MM) method in the ONIOM formalism, the catalytic mechanism of OCD has been examined. The rate limiting step is calculated to be the initial step in the overall mechanism: hydride transfer from the + + L-ornithine’s Cα–H group to the NAD cofactor with concomitant formation of a Cα=NH2 Schiff base with a barrier of 90.6 kJ mol−1. Importantly, no water is observed within the + active site during the MD simulations suitably positioned to hydrolyze the Cα=NH2 intermediate to form the corresponding carbonyl. -

Journal of Biotechnology 193 (2015) 37–40
Journal of Biotechnology 193 (2015) 37–40 Contents lists available at ScienceDirect Journal of Biotechnology j ournal homepage: www.elsevier.com/locate/jbiotec Short communication Blockage of the pyrimidine biosynthetic pathway affects riboflavin production in Ashbya gossypii 1 1 ∗ Rui Silva , Tatiana Q. Aguiar , Lucília Domingues CEB – Centre of Biological Engineering, University of Minho, 4710-057 Braga, Portugal a r t i c l e i n f o a b s t r a c t Article history: The Ashbya gossypii riboflavin biosynthetic pathway and its connection with the purine pathway have Received 10 September 2014 been well studied. However, the outcome of genetic alterations in the pyrimidine pathway on riboflavin Received in revised form 6 November 2014 production by A. gossypii had not yet been assessed. Here, we report that the blockage of the de novo Accepted 7 November 2014 pyrimidine biosynthetic pathway in the recently generated A. gossypii Agura3 uridine/uracil auxotrophic Available online 15 November 2014 strain led to improved riboflavin production on standard agar-solidified complex medium. When extra uridine/uracil was supplied, the production of riboflavin by this auxotroph was repressed. High concen- Keywords: trations of uracil hampered this (and the parent) strain growth, whereas excess uridine favored the A. Ashbya gossypii gossypii Agura3 growth. Considering that the riboflavin and the pyrimidine pathways share the same Pyrimidine pathway precursors and that riboflavin overproduction may be triggered by nutritional stress, we suggest that Riboflavin production Uridine/uracil auxotrophy overproduction of riboflavin by the A. gossypii Agura3 may occur as an outcome of a nutritional stress Nutritional stress response and/or of an increased availability in precursors for riboflavin biosynthesis, due to their reduced consumption by the pyrimidine pathway. -

Electronic Supplementary Material (ESI) for Analyst. This Journal Is © the Royal Society of Chemistry 2017
Electronic Supplementary Material (ESI) for Analyst. This journal is © The Royal Society of Chemistry 2017 Supplemental Table 2. Proteins Increased in Either Blood or Horizon media Table 2A. Proteins Increased in Spores Produced on Horizon Soil Over Spores Produced on Blood Medium quasi.fdr Protein Protein Class/Name KEGG Pathway Names or Function (if ID Pathways found in KEGG) Amino Acid Metabolism bat00250, bat00280, Alanine, aspartate and glutamate metabolism, bat00410, Valine, leucine and isoleucine degradation, bat00640, beta-Alanine metabolism to acetyl CoA, 4.50E-07 BAS0310 4-aminobutyrate aminotransferase bat00650 Propanoate metabolism, Butanoate metabolism bat00270, bat00330, Cysteine and methionine metabolism, Arginine bat00410, and proline metabolism, beta-Alanine 3.84E-10 BAS5060 spermidine synthase bat00480 metabolism, Glutathione metabolism bat00270, bat00330, Cysteine and methionine metabolism, Arginine bat00410, and proline metabolism, beta-Alanine 2.30E-09 BAS5219 spermidine synthase bat00480 metabolism, Glutathione metabolism bat00250, Alanine, aspartate and glutamate metabolism, 6.94E-07 BAS0561 alanine dehydrogenase bat00430 Taurine and hypotaurine metabolism bat00250, Alanine, aspartate and glutamate metabolism, 5.23E-07 BAS4521 alanine dehydrogenase bat00430 Taurine and hypotaurine metabolism 0.005627 BAS5218 agmatinase, putative bat00330 Arginine and proline metabolism 2,3,4,5-tetrahydropyridine-2- carboxylate N-succinyltransferase, 1.40E-10 BAS3891 putative bat00300 Lysine biosynthesis bat00010, bat00020, Glycolysis -

(12) Patent Application Publication (10) Pub. No.: US 2013/0089535 A1 Yamashiro Et Al
US 2013 0089535A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2013/0089535 A1 Yamashiro et al. (43) Pub. Date: Apr. 11, 2013 (54) AGENT FOR REDUCING ACETALDEHYDE Publication Classification NORAL CAVITY (51) Int. Cl. (75) Inventors: Kan Yamashiro, Kakamigahara-shi (JP); A68/66 (2006.01) Takahumi Koyama, Kakamigahara-shi A638/51 (2006.01) (JP) A61O 11/00 (2006.01) A638/44 (2006.01) Assignee: AMANOENZYME INC., Nagoya-shi (52) U.S. Cl. (73) CPC. A61K 8/66 (2013.01); A61K 38/44 (2013.01); (JP) A61 K38/51 (2013.01); A61O II/00 (2013.01) (21) Appl. No.: 13/703,451 USPC .......... 424/94.4; 424/94.5; 435/191: 435/232 (22) PCT Fled: Jun. 7, 2011 (57) ABSTRACT Disclosed herein is a novel enzymatic agent effective in (86) PCT NO.: PCT/UP2011/062991 reducing acetaldehyde in the oral cavity. It has been found S371 (c)(1), that an aldehyde dehydrogenase derived from a microorgan (2), (4) Date: Dec. 11, 2012 ism belonging to the genus Saccharomyces and a threonine aldolase derived from Escherichia coli are effective in reduc (30) Foreign Application Priority Data ing low concentrations of acetaldehyde. Therefore, an agent for reducing acetaldehyde in the oral cavity is provided, Jun. 19, 2010 (JP) ................................. 2010-140O26 which contains these enzymes as active ingredients. Patent Application Publication Apr. 11, 2013 Sheet 1 of 2 US 2013/0089535 A1 FIG 1) 10.5 1 0 9.9.5 8. 5 CONTROL TA AD (BSA) ENZYME Patent Application Publication Apr. 11, 2013 Sheet 2 of 2 US 2013/0089535 A1 FIG 2) 110 the CONTROL (BSA) 100 354. -

Phosphatidylinositol-3-Kinase in Tomato (Solanum Lycopersicum. L) Fruit and Its Role in Ethylene Signal Transduction and Senescence
Phosphatidylinositol-3-Kinase in Tomato (Solanum lycopersicum. L) Fruit and Its Role in Ethylene Signal Transduction and Senescence by Mohd Sabri Pak Dek A Thesis presented to The University of Guelph In partial fulfilment of requirements for the degree of Doctor of Philosophy in Plant Agriculture Guelph, Ontario, Canada © Mohd Sabri Pak Dek,June, 2015 ABSTRACT PHOSPHATIDYLINOSITOL-3-KINASE IN TOMATO (SOLANUM LYCOPERSICUM. L) FRUIT AND ITS ROLE IN ETHYLENE SIGNAL TRANSDUCTION AND SENESCENCE Mohd Sabri Pak Dek Co-Advisors: University of Guelph, 2015 Professor G. Paliyath Professor J. Subramanian The ripening process is initiated by ethylene through a signal transduction cascade leads to the expression of ripening-related genes and catabolism of membrane, cell wall, and storage components. One of the minor components in membrane phospholipids is phosphatidylinositol (PI). Phosphatidylinositol-3-kinase (PIK) is an enzyme that phosphorylates PI at the 3-OH position of inositol head group to produce phosphatidylinositol 3-phosphate (PI3P). Phosphorylation of PI may be an early event in the ethylene signal transduction pathway that generates negatively charged domains on the plasma membrane. PI3P domains may potentially serve as a docking site for phospholipase D (PLD) after ethylene stimulation. It is hypothesized that ethylene stimulation may activate PI3K resulting in enhanced level of phosphorylated phosphatidylinositol. However, the properties and function of PI3K is not well understood in plants. In the present study, the effect of PI3K inhibition during tomato fruit ripening was evaluated. This study demonstrated that PI3K activity is required for normal ripening process of the fruit. Inhibition of PI3K activity using wortmannin significantly reduced tomato ripening process. -

Control Engineering Perspective on Genome-Scale Metabolic Modeling
Control Engineering Perspective on Genome-Scale Metabolic Modeling by Andrew Louis Damiani A dissertation submitted to the Graduate Faculty of Auburn University in partial fulfillment of the requirements for the Degree of Doctor of Philosophy Auburn, Alabama December 12, 2015 Key words: Scheffersomyces stipitis, Flux Balance Analysis, Genome-scale metabolic models, System Identification Framework, Model Validation, Phenotype Phase Plane Analysis Copyright 2015 by Andrew Damiani Approved by Jin Wang, Chair, Associate Professor of Chemical Engineering Q. Peter He, Associate Professor of Chemical Engineering, Tuskegee University Thomas W. Jeffries, Professor of Bacteriology, Emeritus; University of Wisconsin-Madison Allan E. David, Assistant Professor of Chemical Engineering Yoon Y. Lee, Professor of Chemical Engineering Abstract Fossil fuels impart major problems on the global economy and have detrimental effects to the environment, which has caused a world-wide initiative of producing renewable fuels. Lignocellulosic bioethanol for renewable energy has recently gained attention, because it can overcome the limitations that first generation biofuels impose. Nonetheless, in order to have this process commercialized, the biological conversion of pentose sugars, mainly xylose, needs to be improved. Scheffersomyces stipitis has a physiology that makes it a valuable candidate for lignocellulosic bioethanol production, and lately has provided genes for designing recombinant Saccharomyces cerevisiae. In this study, a system biology approach was taken to understand the relationship of the genotype to phenotype, whereby genome-scale metabolic models (GSMMs) are used in conjunction with constraint-based modeling. The major restriction of GSMMs is having an accurate methodology for validation and evaluation. This is due to the size and complexity of the models. -

Protein Expression Profile of Gluconacetobacter Diazotrophicus PAL5, a Sugarcane Endophytic Plant Growth-Promoting Bacterium
Proteomics 2008, 8, 1631–1644 DOI 10.1002/pmic.200700912 1631 RESEARCH ARTICLE Protein expression profile of Gluconacetobacter diazotrophicus PAL5, a sugarcane endophytic plant growth-promoting bacterium Leticia M. S. Lery1, 2, Ana Coelho1, 3, Wanda M. A. von Kruger1, 2, Mayla S. M. Gonc¸alves1, 3, Marise F. Santos1, 4, Richard H. Valente1, 5, Eidy O. Santos1, 3, Surza L. G. Rocha1, 5, Jonas Perales1, 5, Gilberto B. Domont1, 4, Katia R. S. Teixeira1, 6 and Paulo M. Bisch1, 2 1 Rio de Janeiro Proteomics Network, Rio de Janeiro, Brazil 2 Unidade Multidisciplinar de Genômica, Instituto de Biofísica Carlos Chagas Filho, Universidade Federal do Rio de Janeiro, Rio de Janeiro, Brazil 3 Departamento de Genética, Instituto de Biologia, Universidade Federal do Rio de Janeiro, Rio de Janeiro, Brazil 4 Laboratório de Química de Proteínas, Departamento de Bioquímica, Instituto de Química, Universidade Federal do Rio de Janeiro, Rio de Janeiro, Brazil 5 Laboratório de Toxinologia, Departamento de Fisiologia e Farmacodinâmica- Instituto Oswaldo Cruz- Fundac¸ão Oswaldo Cruz, Rio de Janeiro, Rio de Janeiro, Brazil 6 Laboratório de Genética e Bioquímica, Embrapa Agrobiologia, Seropédica, Brazil This is the first broad proteomic description of Gluconacetobacter diazotrophicus, an endophytic Received: September 25, 2007 bacterium, responsible for the major fraction of the atmospheric nitrogen fixed in sugarcane in Revised: December 18, 2007 tropical regions. Proteomic coverage of G. diazotrophicus PAL5 was obtained by two independent Accepted: December 19, 2007 approaches: 2-DE followed by MALDI-TOF or TOF-TOF MS and 1-DE followed by chromatog- raphy in a C18 column online coupled to an ESI-Q-TOF or ESI-IT mass spectrometer. -

The Phylogenetic Extent of Metabolic Enzymes and Pathways José Manuel Peregrin-Alvarez, Sophia Tsoka, Christos A
Downloaded from genome.cshlp.org on October 8, 2021 - Published by Cold Spring Harbor Laboratory Press Letter The Phylogenetic Extent of Metabolic Enzymes and Pathways José Manuel Peregrin-Alvarez, Sophia Tsoka, Christos A. Ouzounis1 Computational Genomics Group, The European Bioinformatics Institute, EMBL Cambridge Outstation, Cambridge CB10 1SD, UK The evolution of metabolic enzymes and pathways has been a subject of intense study for more than half a century. Yet, so far, previous studies have focused on a small number of enzyme families or biochemical pathways. Here, we examine the phylogenetic distribution of the full-known metabolic complement of Escherichia coli, using sequence comparison against taxa-specific databases. Half of the metabolic enzymes have homologs in all domains of life, representing families involved in some of the most fundamental cellular processes. We thus show for the first time and in a comprehensive way that metabolism is conserved at the enzyme level. In addition, our analysis suggests that despite the sequence conservation and the extensive phylogenetic distribution of metabolic enzymes, their groupings into biochemical pathways are much more variable than previously thought. One of the fundamental tenets in molecular biology was ex- reliable source of metabolic information. The EcoCyc data- pressed by Monod, in his famous phrase “What is true for base holds information about the full genome and all known Escherichia coli is true for the elephant” (Jacob 1988). For a metabolic pathways of Escherichia coli (Karp et al. 2000). Re- long time, this statement has inspired generations of molecu- cently, the database has been used to represent computational lar biologists, who have used Bacteria as model organisms to predictions of other organisms (Karp 2001). -

1 Metabolic Dysfunction Is Restricted to the Sciatic Nerve in Experimental
Page 1 of 255 Diabetes Metabolic dysfunction is restricted to the sciatic nerve in experimental diabetic neuropathy Oliver J. Freeman1,2, Richard D. Unwin2,3, Andrew W. Dowsey2,3, Paul Begley2,3, Sumia Ali1, Katherine A. Hollywood2,3, Nitin Rustogi2,3, Rasmus S. Petersen1, Warwick B. Dunn2,3†, Garth J.S. Cooper2,3,4,5* & Natalie J. Gardiner1* 1 Faculty of Life Sciences, University of Manchester, UK 2 Centre for Advanced Discovery and Experimental Therapeutics (CADET), Central Manchester University Hospitals NHS Foundation Trust, Manchester Academic Health Sciences Centre, Manchester, UK 3 Centre for Endocrinology and Diabetes, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, UK 4 School of Biological Sciences, University of Auckland, New Zealand 5 Department of Pharmacology, Medical Sciences Division, University of Oxford, UK † Present address: School of Biosciences, University of Birmingham, UK *Joint corresponding authors: Natalie J. Gardiner and Garth J.S. Cooper Email: [email protected]; [email protected] Address: University of Manchester, AV Hill Building, Oxford Road, Manchester, M13 9PT, United Kingdom Telephone: +44 161 275 5768; +44 161 701 0240 Word count: 4,490 Number of tables: 1, Number of figures: 6 Running title: Metabolic dysfunction in diabetic neuropathy 1 Diabetes Publish Ahead of Print, published online October 15, 2015 Diabetes Page 2 of 255 Abstract High glucose levels in the peripheral nervous system (PNS) have been implicated in the pathogenesis of diabetic neuropathy (DN). However our understanding of the molecular mechanisms which cause the marked distal pathology is incomplete. Here we performed a comprehensive, system-wide analysis of the PNS of a rodent model of DN. -

Genome of Phaeocystis Globosa Virus Pgv-16T Highlights the Common Ancestry of the Largest Known DNA Viruses Infecting Eukaryotes
Genome of Phaeocystis globosa virus PgV-16T highlights the common ancestry of the largest known DNA viruses infecting eukaryotes Sebastien Santinia, Sandra Jeudya, Julia Bartolia, Olivier Poirota, Magali Lescota, Chantal Abergela, Valérie Barbeb, K. Eric Wommackc, Anna A. M. Noordeloosd, Corina P. D. Brussaardd,e,1, and Jean-Michel Claveriea,f,1 aStructural and Genomic Information Laboratory, Unité Mixte de Recherche 7256, Centre National de la Recherche Scientifique, Aix-Marseille Université, 13288 Marseille Cedex 9, France; bCommissariat à l’Energie Atomique–Institut de Génomique, 91057 Evry Cedex, France; cDepartment of Plant and Soil Sciences, University of Delaware, Newark, DE 19711; dDepartment of Biological Oceanography, Royal Netherlands Institute for Sea Research, NL-1790 AB Den Burg (Texel), The Netherlands; eAquatic Microbiology, Institute for Biodiversity and Ecosystem Dynamics, University of Amsterdam, Amsterdam, The Netherlands; and fService de Santé Publique et d’Information Médicale, Hôpital de la Timone, Assistance Publique–Hôpitaux de Marseille, FR-13385 Marseille, France Edited by James L. Van Etten, University of Nebraska, Lincoln, NE, and approved May 1, 2013 (received for review February 22, 2013) Large dsDNA viruses are involved in the population control of many viruses: 730 kb and 1.28 Mb for CroV and Megavirus chilensis, globally distributed species of eukaryotic phytoplankton and have respectively. Other studies, targeting virus-specific genes [e.g., a prominent role in bloom termination. The genus Phaeocystis (Hap- DNA polymerase B (8) or capsid proteins (9)] have suggested tophyta, Prymnesiophyceae) includes several high-biomass-forming a close phylogenetic relationship between Mimivirus and several phytoplankton species, such as Phaeocystis globosa, the blooms of giant dsDNA viruses infecting various unicellular algae such as which occur mostly in the coastal zone of the North Atlantic and the Pyramimonas orientalis (Chlorophyta, Prasinophyceae), Phaeocys- North Sea.