Nature Medicine Essay
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Hypertension, Cardiac Hypertrophy, and Sudden Death in Mice Lacking
Proc. Natl. Acad. Sci. USA Vol. 94, pp. 14730–14735, December 1997 Medical Sciences Hypertension, cardiac hypertrophy, and sudden death in mice lacking natriuretic peptide receptor A (gene targetingyechocardiographyycardiac dilatationyinterstitial fibrosisyaortic dissection) PAULA M. OLIVER*, JENNIFER E. FOX*, RON KIM*, HOWARD A. ROCKMAN†,HYUNG-SUK KIM*, i ROBERT L. REDDICK‡,KAILASH N. PANDEY§,SHARON L. MILGRAM¶,OLIVER SMITHIES*, AND NOBUYO MAEDA* *Department of Pathology, University of North Carolina, 710 Brinkhous-Bullitt Building, Chapel Hill, NC 27599-7525; Departments of †Medicine and ¶Physiology, University of North Carolina, Chapel Hill, NC 27599; ‡Department of Pathology, The University of Texas Health Science Center, 7703 Floyd Curl Drive, San Antonio, TX 78284; and §Department of Biochemistry and Molecular Biology, Medical College of Georgia, Augusta, GA 30912 Contributed by Oliver Smithies, October 27, 1997 ABSTRACT Natriuretic peptides, produced in the heart, of blood pressure and in the cardiovascular response to bind to the natriuretic peptide receptor A (NPRA) and cause sustained hypertension we have made mice completely lacking vasodilation and natriuresis important in the regulation of this receptor. Our results demonstrate that NPRA deficiency blood pressure. We here report that mice lacking a functional in mice leads to elevated blood pressures and, particularly in Npr1 gene coding for NPRA have elevated blood pressures and males, to cardiac hypertrophy and sudden death. hearts exhibiting marked hypertrophy with interstitial fibro- sis resembling that seen in human hypertensive heart disease. Echocardiographic evaluation of the mice demonstrated a MATERIALS AND METHODS compensated state of systemic hypertension in which cardiac Gene Targeting. Three electroporations of embryonic stem hypertrophy and dilatation are evident but with no reduction (ES) cells (E14TG2a) were carried out as described (11). -

Apolipoprotein E Isoforms in Diabetic Dyslipidemia and Atherosclerosis
Apolipoprotein E isoforms in diabetic dyslipidemia and atherosclerosis Lance Johnson Diabetic Dyslipidemia Cluster of harmful changes to lipoprotein metabolism commonly noted in diabetic patients Characterized by high VLDL triglycerides, low HDL-cholesterol, and a increase in small, dense LDL particles Increases the risk of developing atherosclerosis Apolipoprotein E Isoforms ApoE2 ApoE3 ApoE4 position 112 cys cys arg position 158 cys arg arg frequency 7.2% 78.3% 14.3% LDLR affinity <1% 100% >100% plasma TC -6% ref. +6% MI risk reduced ref. Increased (type III) Plasma lipoproteins Normal LDLR Increased LDLR 2m (350mg/dl) 2h (100mg/dl) 60 VLDL HDL Mice with 40 On a HFW LDL Human apoE2 HDL diet Atherosclerosis No plaques 20 (poor ligand) Cholesterol VLDL LDL 0 10 20 30 10 20 30 60 3m (100mg/dl) 3h (60mg/dl) Mice with HDL 40 Human apoE3 No plaques HDL No plaques (reference) 20 VLDL Cholesterol VLDL 0 10 20 30 10 20 30 4m (100mg/dl) 4h (250mg/dl) 60 HDL VLDL Mice with 40 Human apoE4 No plaques Atherosclerosis HDL 20 (better ligand) Cholesterol VLDL 0 10 20 30 10 20 30 ApoE -Trapping by LDL receptor Figure 2 ApoE trapping. Efficient transfer of apoE4 facilitates internalization of large lipoproteins in 4m mice. In 4h mice, the efficiency of transfer is reduced because apoE4 are trapped by the increased LDLR. Akita-diabetic mice with human apoE isoforms Survival With normal LDLR With human LDLR Diabetic Human ApoE Mice: Survival Rate 110 100 4mAE4 3hA 90 80 3mAE3 2hA 70 60 50 40 30 2mAE2 Percent survival 4hA 20 10 0 0 1 2 3 4 5 6 7 Months of age 800 600 400 Glucose 200 0 2m 2h 3m 3h 4m 4h 200 2mAkita 4h-Akita 150 100 Cholesterol 50 0 2m 2h 3m 3h 4m 4h 150 100 50 Triglycerides 0 2m 2h 3m 3h 4m 4h Two of two -6mo old 4hAkita mice on a low cholesterol diet had atherosclerosis despite of low plasma cholesterol (<120 mg/dl) levels This is the first mouse model of atherosclerosis for which diabetes is required. -

Original Article -Lipoic Acid Prevents the Increase in Atherosclerosis Induced By
Original Article ␣-Lipoic Acid Prevents the Increase in Atherosclerosis Induced by Diabetes in Apolipoprotein E–Deficient Mice Fed High-Fat/Low-Cholesterol Diet Xianwen Yi and Nobuyo Maeda Considerable evidence indicates that hyperglycemia in- LDL and other lipids by free radicals is one of the most creases oxidative stress and contributes to the increased important factors for the initiation of atherosclerosis (5,6). incidence of atherosclerosis and cardiovascular complica- Oxidative stress also facilitates endothelial cell dysfunc- tions in diabetic patients. To examine the effect of ␣-lipoic tion (7). However, evidence linking antioxidant vitamins to acid, a potent natural antioxidant, on atherosclerosis in diabetes complications in humans is still inconclusive. For diabetic mice, 3-month-old apolipoprotein (apo) E-defi- cient (apoE؊/؊) mice were made diabetic by administering example, although treatment with a high dose of vitamin E streptozotocin (STZ). At 4 weeks after starting the STZ (1,800 IU/day) appeared to be effective in normalizing administration, a high-fat diet with or without ␣-lipoic acid retinal hemodynamic abnormalities and improving renal (1.65 g/kg) was given to the mice and to nondiabetic function in type 1 diabetes in a small randomized trial (8), apoE؊/؊ controls. At 20 weeks, markers of oxidative stress vitamin E at a dose of 400 IU/day had no effect on ؊/؊ were significantly lower in both the diabetic apoE mice cardiovascular outcomes or nephropathy in high-risk pa- ␣ ؊/؊ and their nondiabetic apoE controls with -lipoic acid tients with diabetes, as reported in the HOPE (Heart supplement than in those without it. Remarkably, ␣-lipoic acid completely prevented the increase in plasma total Outcomes Prevention Evaluation) study and MICRO-HOPE cholesterol, atherosclerotic lesions, and the general dete- (Microalbuminuria, Cardiovascular, and Renal Outcomes- rioration of health caused by diabetes. -

Hypertension and Abnormal Fat Distribution but Not Insulin Resistance in Mice with P465L Pparγ
Hypertension and abnormal fat distribution but not insulin resistance in mice with P465L PPARγ Yau-Sheng Tsai, … , Jason K. Kim, Nobuyo Maeda J Clin Invest. 2004;114(2):240-249. https://doi.org/10.1172/JCI20964. Article Cardiology Peroxisome proliferator–activated receptor γ (PPARγ), the molecular target of a class of insulin sensitizers, regulates adipocyte differentiation and lipid metabolism. A dominant negative P467L mutation in the ligand-binding domain of PPARγ in humans is associated with severe insulin resistance and hypertension. Homozygous mice with the equivalent P465L mutation die in utero. Heterozygous mice grow normally and have normal total adipose tissue weight. However, they have reduced interscapular brown adipose tissue and intra-abdominal fat mass, and increased extra-abdominal subcutaneous fat, compared with wild-type mice. They have normal plasma glucose levels and insulin sensitivity, and increased glucose tolerance. However, during high-fat feeding, their plasma insulin levels are mildly elevated in association with a significant increase in pancreatic islet mass. They are hypertensive, and expression of the angiotensinogen gene is increased in their subcutaneous adipose tissues. The effects of P465L on blood pressure, fat distribution, and insulin sensitivity are the same in both male and female mice regardless of diet and age. Thus the P465L mutation alone is sufficient to cause abnormal fat distribution and hypertension but not insulin resistance in mice. These results provide genetic evidence for a critical role for PPARγ in blood pressure regulation that is not dependent on altered insulin sensitivity. Find the latest version: https://jci.me/20964/pdf Research article Related Commentary, page 163 Hypertension and abnormal fat distribution but not insulin resistance in mice with P465L PPARγ Yau-Sheng Tsai,1 Hyo-Jeong Kim,2 Nobuyuki Takahashi,1 Hyung-Suk Kim,1 John R. -

Professor Oliver Smithies Was Born on June 23, 1925, in Halifax, England, and As a Child Went to a Small School in Copley Village Near to Halifax
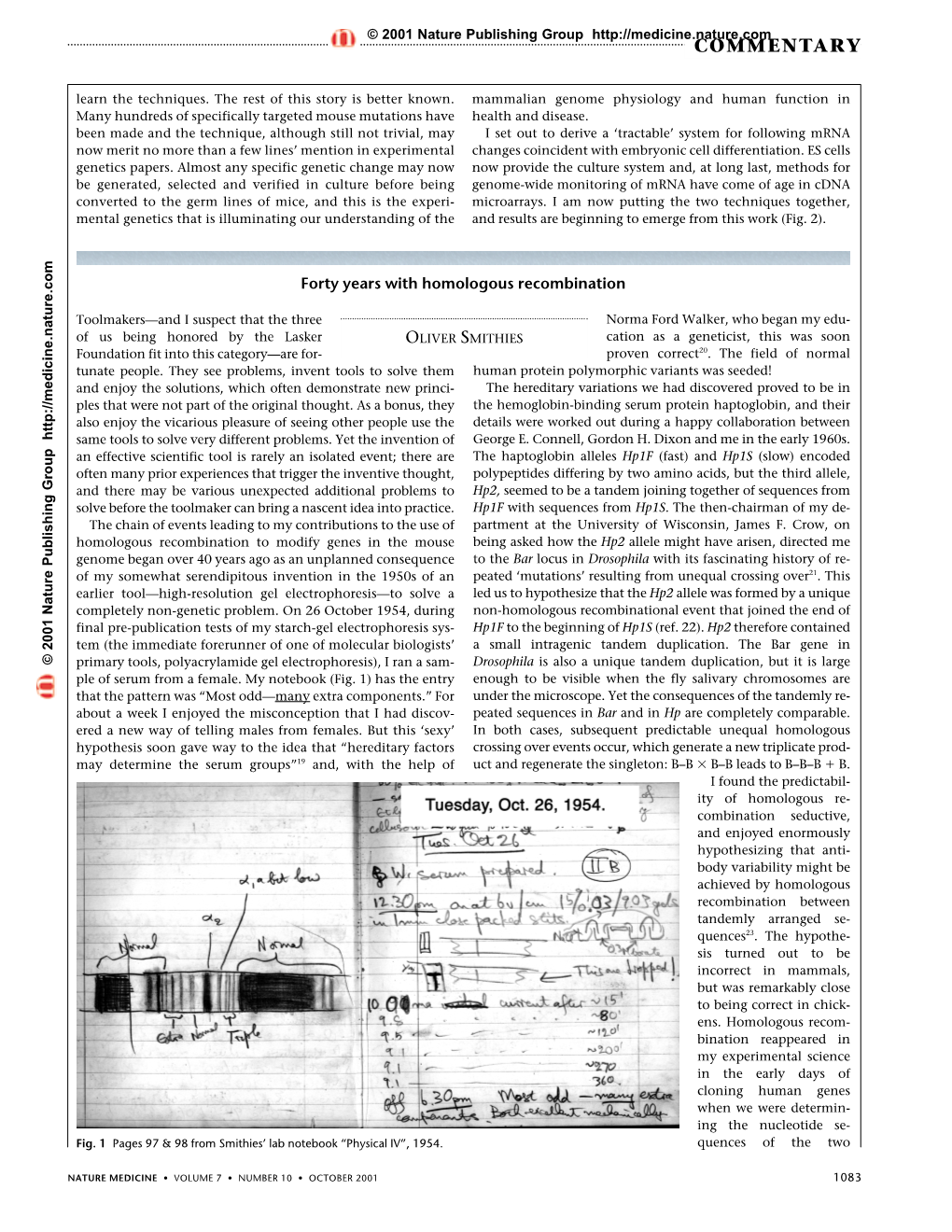
Professor Oliver Smithies was born on June 23, 1925, in Halifax, England, and as a child went to a small school in Copley village near to Halifax. Subsequently he was a student at Heath Grammar School in Halifax until he was awarded a scholarship from Balliol College, Oxford University. At Oxford he received a Bachelor of Arts Degree in Physiology with First Class Honors in 1946. In 1951, he obtained his M.A. and D.Phil. degrees in Biochemistry from Oxford. Professor Smithies then moved to the United States as a Postdoctoral Fellow in Physical Chemistry at the University of Wisconsin. After two years at Wisconsin, Professor Smithies accepted a position at the Connaught Medical Research Laboratories in Toronto and stayed there from 1953 to 1960, first as a Research Assistant and then as a Research Associate. In 1960, Professor Smithies returned to Wisconsin as Assistant Professor of Medical Genetics and Genetics, advancing to Full Professor by 1963. He was named the Leon J. Cole Professor in 1971. In 1988, he joined the Department of Pathology at the University of North Carolina, as the Excellence Professor of Pathology. He is now the Weatherspoon Eminent Distinguished Professor of Pathology and Laboratory Medicine at UNC, and remains an actively engaged scientist working at the bench in the laboratory he shares with his wife, Nobuyo Maeda who is the Robert H. Wagner Distinguished Professor of Pathology and Laboratory Medicine. In the mid-1950s, Professor Smithies described the first high resolution electrophoresis system (starch gel), and with it he discovered that normal humans have unsuspected inherited differences in their proteins. -

In 1996, I Graduated from the Department of Biochemistry In
Designer mice for human disease - A close view of Nobel Laureate: Oliver Smithies Yau-Sheng Tsai, Institute of Clinical Medicine, National Cheng Kung University, Tainan, Taiwan Pei-Jane Tsai, National Laboratory Animal Center, National Applied Research Laboratories, Taipei, Taiwan Man-Jin Jiang, Cardiovascular Research Center, National Cheng Kung University, Tainan, Taiwan Cherng-Shyang Chang, Institute of Basic Medical Sciences, National Cheng Kung University, Tainan, Taiwan Two major innovations in the 1980s have changed all fields of biomedicine. Combination of gene targeting in mammalian cells with culture of embryonic stem cell enables scientists to study specific genes by creating “knockout mice”. Through targeting and removing a specific gene, researchers can find out what happens once it is gone! Working on gene targeting in the Laboratory of Oliver Smithies and his wife, Nobuyo Maeda, in 2000~2006 made my American journey enlightening and fulfilling. Things I learned from Oliver and Nobuyo are not only research, but also the scientific temper and attitude these two distinguished scientists represent. Designer mice for human disease The 2007 Nobel Prize in Physiology or Medicine goes to a series of ground breaking discoveries regarding the gene targeting in mice, commonly called knockout technology. The principles for these discoveries are to introduce specific gene modification in mice by the use of embryonic stem (ES) cell (Figure 1). Drs. Mario R. Capecchi of the University of Utah’s Howard Hughes Medical Institute, Martin J. Evans of the Cardiff University in United Kingdom, and Oliver Smithies of the University of North Carolina at Chapel Hill will share this year’s Nobel Prize. -

Maeda, Nobuyo (2011)
AMDCC Annual Report (2011) PI: MAEDA, NOBUYO Project Title: Dyslipidemia, Lipoic Acid and Diabetic Vascular Complications in Humanized Mice Grant Number: U01 HL087946 Abstract: While diabetes mellitus can lead to serious damage to many organs, cardiovascular diseases are the major cause of death and morbidity in diabetic patients. Overall, patients with diabetes have a three to five fold increased risk of coronary artery diseases compared to non-diabetics. Our goal is to use mouse genetics for identifying genetic risk factors for the vascular complications of diabetes and for unraveling underlying mechanisms. Although a significant increase in atherosclerosis by diabetes has been demonstrated in atherogenic mouse models, none of these mouse models faithfully replicates the types of dyslipidemia associated with diabetes in humans. We postulate that this failure is due to differences in the relative levels of plasma low density lipoprotein (LDL) and plasma high density lipoprotein (HDL) that are controlled by genetic differences between the two species and genetic polymorphisms in humans. Thus our first hypothesis is that humanizing genes that are involved in lipoprotein metabolism in mice so that they develop a more human-like diabetic dyslipidemia will cause them to replicate better the cardiovascular problems of human diabetic patients. We will test this hypothesis in Specific Aim 1 by inducing diabetes in mice with humanized apoE of the three isoforms (E2, E3, and E4) and humanized LDL receptor (LDLR), with or without overexpression of human apoB. We predict that this will lead to diabetic dyslipidemia and accelerated atherosclerosis in an apoE isoform dependent manner. Our second hypothesis is that since diabetes is generally acknowledged to induce oxidative stress, genetically determined differences in the levels of endogenous anti-oxidants affect the development of cardiovascular complications,. -

Molecular and Cellular Mediators in Radiation-Induced Lung
MOLECULAR AND CELLULAR MEDIATORS IN RADIATION-INDUCED LUNG INJURY Xuebin Yang A dissertation submitted to the faculty of the University of North Carolina at Chapel Hill in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Department of Pathology and Laboratory Medicine. Chapel Hill 2009 Approved by: Suzanne Kirby, M.D. Ph.D. Donald N. Cook, Ph.D. Virginia L. Godfrey, DVM. Ph.D. Nobuyo N. Maeda, Ph.D. Stephen Tilley, M.D. Abstract Xuebin Yang: Molecular and Cellular Mediators In Radiation-Induced Lung Injury (Under the direction of Suzanne Kirby, M.D. Ph.D) Radiation-induced lung injury is a common adverse effect in patients receiving thoracic irradiation and for which, there is currently no effective therapy. Using a murine model of thoracic irradiation, we found that mice deficient in either the chemokine CCL3 or one of its receptors, CCR1, are significantly protected from radiation lung injury. This protected phenotype includes improved survival, virtually no pneumonitis or fibrosis, and preserved lung function when compared to wild-type mice. We further showed that a specific CCR1 inhibitor, BX471 provided similar protection. Therefore, CCR1 is a promising target for reducing radiation lung injury. To investigate the mechanisms by which CCL3/CCR1 signalling mediates radiation lung injury, we evaluated their influence on lung inflammation after irradiation. When compared with irradiated WT mice, irradiated CCL3- and CCR1-deficient mice had less lung + + infiltration of CD4 and CD8 T cells; however, CD4-deficient mice showed only partial protection, while CD8-deficient mice had slightly worse fibrosis. We further analyzed + inflammatory cytokines and different subsets of CD4 lymphocytes, T 1, T 2, T 17 and H H H + Treg cells, in our model. -

NIH Public Access Author Manuscript Hum Genet
NIH Public Access Author Manuscript Hum Genet. Author manuscript; available in PMC 2015 February 09. NIH-PA Author ManuscriptPublished NIH-PA Author Manuscript in final edited NIH-PA Author Manuscript form as: Hum Genet. 2003 January ; 112(1): 62–70. doi:10.1007/s00439-002-0834-z. Common variations in noncoding regions of the human natriuretic peptide receptor A gene have quantitative effects Joshua W. Knowles, Department of Pathology and Laboratory Medicine, University of North Carolina, Chapel Hill, NC 27599–7525, USA Curriculum in Genetics and Molecular Biology, University of North Carolina, Chapel Hill, NC 27599–7525, USA Laurie M. Erickson, Department of Pathology and Laboratory Medicine, University of North Carolina, Chapel Hill, NC 27599–7525, USA Vanessa K. Guy, Department of Pathology and Laboratory Medicine, University of North Carolina, Chapel Hill, NC 27599–7525, USA Carlie S. Sigel, Department of Pathology and Laboratory Medicine, University of North Carolina, Chapel Hill, NC 27599–7525, USA Jennifer C. Wilder, and Department of Pathology and Laboratory Medicine, University of North Carolina, Chapel Hill, NC 27599–7525, USA Nobuyo Maeda Department of Pathology and Laboratory Medicine, University of North Carolina, Chapel Hill, NC 27599–7525, USA [email protected], Tel.: +1-919-9666912, Fax: +1-919-9668800 Curriculum in Genetics and Molecular Biology, University of North Carolina, Chapel Hill, NC 27599–7525, USA Abstract Genetic susceptibility to common conditions, such as essential hypertension and cardiac hypertrophy, is probably determined by various combinations of small quantitative changes in the expression of many genes. NPR1, coding for natriuretic peptide receptor A (NPRA), is a potential candidate, because NPRA mediates natriuretic, diuretic, and vasorelaxing actions of the nariuretic peptides, and because genetically determined quantitative changes in the expression of this gene affect blood pressure and heart weight in a dose-dependent manner in mice. -

Maeda, Nobuyo (2009)
Animal Models of Diabetic Complications Consortium (U01 HL087946) Annual Report (2009) “Dislipidemia, Lipoic Acid and Diabetic Vascular Complications in Humanized Mice” The University of North Carolina at Chapel Hill Department of Pathology and Laboratory Medicine Principal Investigator Nobuyo Maeda Address: 710 Brinkhous-Bullitt Building, CB7527, Chapel Hill NC 27599-7525 Phone: 919-966-6914 E-mail: [email protected] 1 Table of Contents Page Part A: Principal Investigator’s Summary 3 1. Project Accomplishments (2009) 4 2. Collaboration 7 3. Address previous EAC comments 8 4. Publications 9 Part B: Individual Project Reports by Responsible Investigator (if applicable) NA Project 1 (if applicable): “Title” X Responsible Investigator: Name Project 2 (if applicable): “Title” X Responsible Investigator: Name Project 3 (if applicable): “Title” X Responsible Investigator: Name 2 Animal Models of Diabetic Complications Consortium (U01 HL087946) Part A: Principal Investigator’s Summary 3 1. Program Accomplishments: Hypothesis 1. Mice humanized lipoprotein metabolism system will develop a more human-like diabetic dyslipidemia and cardiovascular problems. 2. Genetically determined differences in the levels of endogenous antioxidants affect the development of cardiovascular complications of diabetes. Recent Progress and Major Accomplishments 1-1. Akita mice expressing human apoE4 and human LDLR develop atherosclerosis without severe hypercholesterolemia. Non-diabetic E3 and E4 mice with 2 fold normal expression of the human LDLR (3h and 4h mice, respectively) have similar plasma fasting glucose and triglyceride levels to wild type mice. Their cholesterol levels are reduced because of reduced HDL-cholesterol to one third when on a normal chow (low-fat diet), and neither of these strains develops atherosclerosis. -

Spontaneous Hypercholesterolemia and Arterial Lesions in Mice Lacking Apolipoprotein E SH Zhang, RL Reddick, JA Piedrahita and N Maeda
.! , ......... 1.. .., ., ------ ..ll, - .:U, . 1 4. M-1 W1411.'AN.4"lW*v,1 or,-Al11 m-, R a.. .....' the refolding of the short-time denatured 3. R. J. Ellis and S. M. van der Vies, Annu. Rev. 19. H. R. B. Pelham, Cell 46, 959 (1986). Biochem. 60, 321 (1991). 20. G. C. Flynn, J. Pohl, M. T. Flocco, J. E. Rothman, enzyme, but that a fraction of the prolines 4. F. X. Schmid, Curr. Opin. Struct. Biol. 1, 36 (1991). Nature 353, 726 (1991). must be very rapidly buried during the 5. G. Fischer and F. X. Schmid, Biochemistry 29, 21. P.-0. FreskgArd, U. Carlsson, L. G. MArtensson, initiation of the refolding process (pB in 2205 (1990). B.-H. Jonsson, FEBS Lett. 289, 117 (1991). 6. A. E. Eriksson, T. A. Jones, A. Uljas, Proteins 22. R. G. Khalifah, D. J. Strader, S. H. Bryant, S. M. Fig. 3) and thus become inaccessible to Struct. Funct. Genet. 4, 274 (1988). Gibson, Biochemistry 16, 2241 (1977). PPIase. It has been reported (10-12) that 7. C. Fransson et al., FEBS Lett. 296, 90 (1992). 23. L. E. Henderson and D. Henriksson, Anal. Bio- an initiation structure of carbonic anhy- 8. J. L. Cleland and D. I. C. Wang, Biochemistry 29, chem. 51, 288 (1973). 11072 (1990). 24. G. Fischer, H. Bang, C. Mech, Biomed. Biochim. drase is rapidly formed during refolding. 9. J. Buchner et al., ibid. 30, 1586 (1991). Acta 43, 1101 (1984). When the prolines in this core of the 10. U. Carlsson, R. Aasa, L. E. -

Apolipoprotein AI Transgene Corrects Apolipoprotein E Deficiency-Induced Atherosclerosis in Mice
Apolipoprotein AI transgene corrects apolipoprotein E deficiency-induced atherosclerosis in mice. C Pászty, … , J Verstuyft, E M Rubin J Clin Invest. 1994;94(2):899-903. https://doi.org/10.1172/JCI117412. Research Article Apolipoprotein E (apo E)-deficient mice are severely hypercholesterolemic and develop advanced atheromas independent of diet. The C57BL/6 strain differs from most inbred strains by having lower HDL concentrations and a high risk of developing early atherosclerotic lesions when fed an atherogenic diet. The relative HDL deficiency and atherosclerosis susceptibility of the C57BL/6 strain are corrected with the expression of a human apolipoprotein AI (apo AI) transgene in this genetic background. To examine if increases in apo AI and HDL are also effective in minimizing apo E deficiency-- induced atherosclerosis, we introduced the human apo AI transgene into the hypercholesterolemic apo E knockout background. Similar elevations of total plasma cholesterol occurred in both the apo E knockout and apo E knockout mice also expressing the human apo AI transgene. The latter animals, however, also showed a two- to threefold increase in HDL and a sixfold decrease in susceptibility to atherosclerosis. This study demonstrates that elevating the concentration of apo AI reduces atherosclerosis in apo E deficient-mice and suggests that elevation of apo AI and HDL may prove to be a useful approach for treating unrelated causes of heightened atherosclerosis susceptibility. Find the latest version: https://jci.me/117412/pdf Rapid Publication