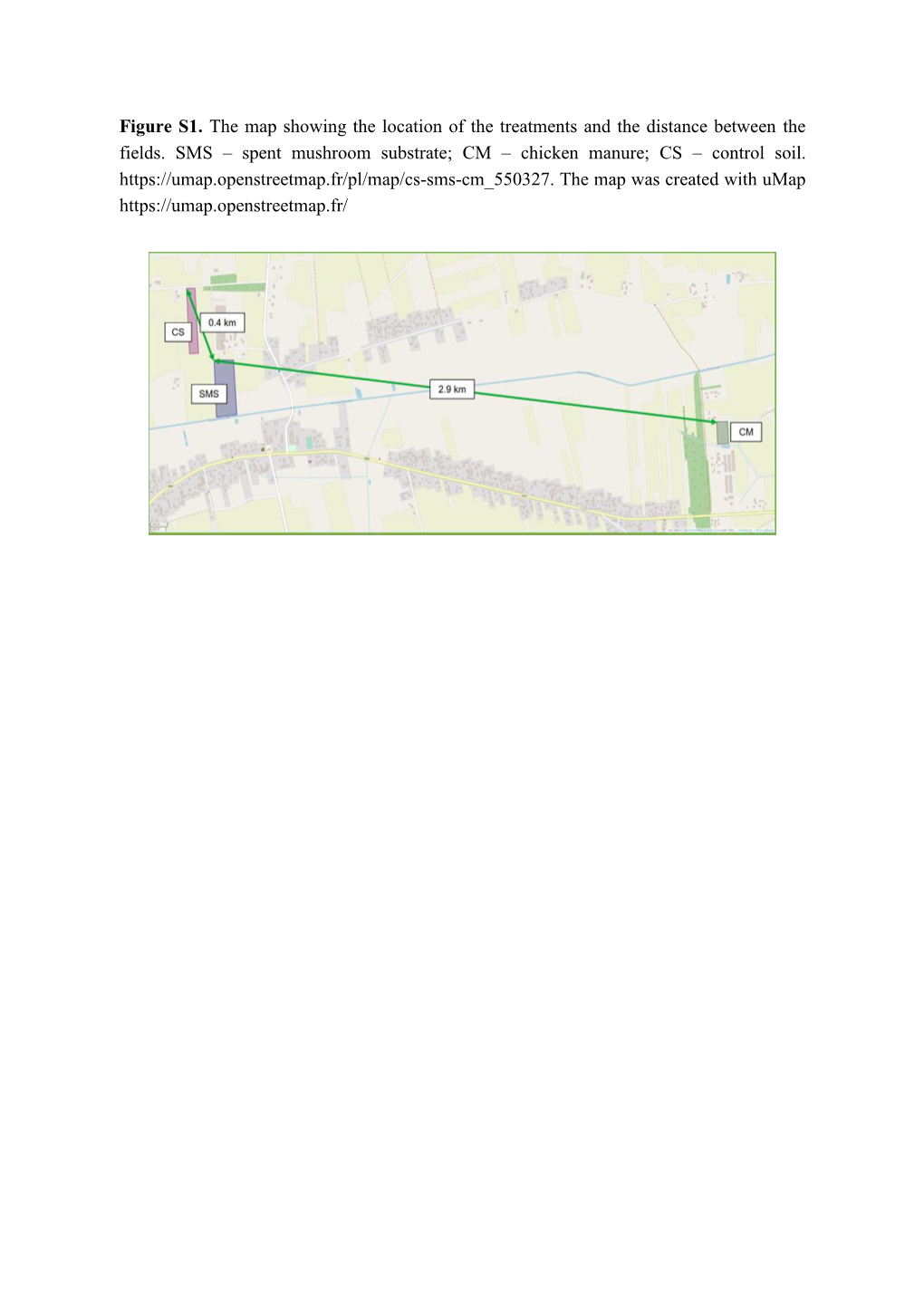
Spent Mushroom Substrate; CM – Chicken Manure; CS – Control Soil
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Microsporum Canis Genesig Standard
Primerdesign TM Ltd Microsporum canis PQ-loop repeat protein gene genesig® Standard Kit 150 tests For general laboratory and research use only Quantification of Microsporum canis genomes. 1 genesig Standard kit handbook HB10.04.10 Published Date: 09/11/2018 Introduction to Microsporum canis Microsporum canis is a zoophilic dermatophyte which is responsible for dermatophytosis in dogs and cats. They cause superficial infections of the scalp (tinea capitis) in humans and ringworm in cats and dogs. They belong to the family Arthrodermataceae and are most commonly found in humid and warm climates. They have numerous multi-celled macroconidia which are typically spindle-shaped with 5-15 cells, verrucose, thick-walled, often having a terminal knob and 35-110 by 12-25 µm. In addition, they produce septate hyphae and microconidia and the Microsporum canis genome is estimated at 23 Mb. The fungus is transmitted from animals to humans when handling infected animals or by contact with arthrospores contaminating the environment. Spores are very resistant and can live up to two years infecting animals and humans. They will attach to the skin and germinate producing hyphae, which will then grow in the dead, superficial layers of the skin, hair or nails. They secrete a 31.5 kDa keratinolytic subtilisin-like protease as well as three other subtilisin- like proteases (SUBs), SUB1, SUB2 and SUB3, which cause damage to the skin and hair follicle. Keratinolytic protease also provides the fungus nutrients by degrading keratin structures into easily absorbable metabolites. Infection leads to a hypersensitive reaction of the skin. The skin becomes inflamed causing the fungus to move away from the site to normal, uninfected skin. -

Diversification of Fungal Chitinases and Their Functional Differentiation in 2 Histoplasma Capsulatum 3
bioRxiv preprint doi: https://doi.org/10.1101/2020.06.09.137125; this version posted June 16, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-ND 4.0 International license. 1 Diversification of fungal chitinases and their functional differentiation in 2 Histoplasma capsulatum 3 4 Kristie D. Goughenour1*, Janice Whalin1, 5 Jason C. Slot2, Chad A. Rappleye1# 6 7 1 Department of Microbiology, Ohio State University 8 2 Department of Plant Pathology, Ohio State University 9 10 11 #corresponding author: 12 [email protected] 13 614-247-2718 14 15 *current affiliation: 16 Division of Pulmonary and Critical Care Medicine 17 University of Michigan 18 VA Ann Arbor Healthcare System, Research Service 19 Ann Arbor, Michigan, USA 20 21 22 running title: Fungal chitinases 23 24 keywords: chitinase, GH18, fungi, Histoplasma 25 bioRxiv preprint doi: https://doi.org/10.1101/2020.06.09.137125; this version posted June 16, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-ND 4.0 International license. 26 ABSTRACT 27 Chitinases enzymatically hydrolyze chitin, a highly abundant biomolecule with many potential 28 industrial and medical uses in addition to their natural biological roles. Fungi are a rich source of 29 chitinases, however the phylogenetic and functional diversity of fungal chitinases are not well 30 understood. -

Fungal Evolution: Major Ecological Adaptations and Evolutionary Transitions
Biol. Rev. (2019), pp. 000–000. 1 doi: 10.1111/brv.12510 Fungal evolution: major ecological adaptations and evolutionary transitions Miguel A. Naranjo-Ortiz1 and Toni Gabaldon´ 1,2,3∗ 1Department of Genomics and Bioinformatics, Centre for Genomic Regulation (CRG), The Barcelona Institute of Science and Technology, Dr. Aiguader 88, Barcelona 08003, Spain 2 Department of Experimental and Health Sciences, Universitat Pompeu Fabra (UPF), 08003 Barcelona, Spain 3ICREA, Pg. Lluís Companys 23, 08010 Barcelona, Spain ABSTRACT Fungi are a highly diverse group of heterotrophic eukaryotes characterized by the absence of phagotrophy and the presence of a chitinous cell wall. While unicellular fungi are far from rare, part of the evolutionary success of the group resides in their ability to grow indefinitely as a cylindrical multinucleated cell (hypha). Armed with these morphological traits and with an extremely high metabolical diversity, fungi have conquered numerous ecological niches and have shaped a whole world of interactions with other living organisms. Herein we survey the main evolutionary and ecological processes that have guided fungal diversity. We will first review the ecology and evolution of the zoosporic lineages and the process of terrestrialization, as one of the major evolutionary transitions in this kingdom. Several plausible scenarios have been proposed for fungal terrestralization and we here propose a new scenario, which considers icy environments as a transitory niche between water and emerged land. We then focus on exploring the main ecological relationships of Fungi with other organisms (other fungi, protozoans, animals and plants), as well as the origin of adaptations to certain specialized ecological niches within the group (lichens, black fungi and yeasts). -

The Soil Fungal Community of Native Woodland in Andean Patagonian
Forest Ecology and Management 461 (2020) 117955 Contents lists available at ScienceDirect Forest Ecology and Management journal homepage: www.elsevier.com/locate/foreco The soil fungal community of native woodland in Andean Patagonian forest: T A case study considering experimental forest management and seasonal effects ⁎ Ayelen Inés Carrona,b, , Lucas Alejandro Garibaldic, Sebastian Marquezd, Sonia Fontenlaa,b a Laboratorio de Microbiología Aplicada y Biotecnología Vegetal y del Suelo, Centro Regional Universitario Bariloche, Universidad Nacional del Comahue (UNComahue), Argentina b Instituto Andino Patagónico de Tecnologías Biológicas y Geoambientales (IPATEC) UNComahue – Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), Argentina c Instituto de Investigaciones en Recursos Naturales, Agroecología y Desarrollo Rural (IRNAD), Sede Andina, Universidad Nacional de Río Negro (UNRN) and CONICET, Argentina d Instituto de Investigación en Biodiversidad y Medio Ambiente (INIBIOMA) UNComahue – Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), Argentina ARTICLE INFO ABSTRACT Keywords: Forest management can alter soil fungal communities which are important in the regulation of biogeochemical Soil fungal classification cycles and other ecosystem services. The current challenge of sustainable management is that management be Diversity analysis carried out while preserving the bioecological aspects of ecosystems. Mixed Patagonian woodlands are subject to Shrubland management continuous disturbance (fire, wood -

Fungal Planet Description Sheets: 716–784 By: P.W
Fungal Planet description sheets: 716–784 By: P.W. Crous, M.J. Wingfield, T.I. Burgess, G.E.St.J. Hardy, J. Gené, J. Guarro, I.G. Baseia, D. García, L.F.P. Gusmão, C.M. Souza-Motta, R. Thangavel, S. Adamčík, A. Barili, C.W. Barnes, J.D.P. Bezerra, J.J. Bordallo, J.F. Cano-Lira, R.J.V. de Oliveira, E. Ercole, V. Hubka, I. Iturrieta-González, A. Kubátová, M.P. Martín, P.-A. Moreau, A. Morte, M.E. Ordoñez, A. Rodríguez, A.M. Stchigel, A. Vizzini, J. Abdollahzadeh, V.P. Abreu, K. Adamčíková, G.M.R. Albuquerque, A.V. Alexandrova, E. Álvarez Duarte, C. Armstrong-Cho, S. Banniza, R.N. Barbosa, J.-M. Bellanger, J.L. Bezerra, T.S. Cabral, M. Caboň, E. Caicedo, T. Cantillo, A.J. Carnegie, L.T. Carmo, R.F. Castañeda-Ruiz, C.R. Clement, A. Čmoková, L.B. Conceição, R.H.S.F. Cruz, U. Damm, B.D.B. da Silva, G.A. da Silva, R.M.F. da Silva, A.L.C.M. de A. Santiago, L.F. de Oliveira, C.A.F. de Souza, F. Déniel, B. Dima, G. Dong, J. Edwards, C.R. Félix, J. Fournier, T.B. Gibertoni, K. Hosaka, T. Iturriaga, M. Jadan, J.-L. Jany, Ž. Jurjević, M. Kolařík, I. Kušan, M.F. Landell, T.R. Leite Cordeiro, D.X. Lima, M. Loizides, S. Luo, A.R. Machado, H. Madrid, O.M.C. Magalhães, P. Marinho, N. Matočec, A. Mešić, A.N. Miller, O.V. Morozova, R.P. Neves, K. Nonaka, A. Nováková, N.H. -

An Evolving Phylogenetically Based Taxonomy of Lichens and Allied Fungi
Opuscula Philolichenum, 11: 4-10. 2012. *pdf available online 3January2012 via (http://sweetgum.nybg.org/philolichenum/) An evolving phylogenetically based taxonomy of lichens and allied fungi 1 BRENDAN P. HODKINSON ABSTRACT. – A taxonomic scheme for lichens and allied fungi that synthesizes scientific knowledge from a variety of sources is presented. The system put forth here is intended both (1) to provide a skeletal outline of the lichens and allied fungi that can be used as a provisional filing and databasing scheme by lichen herbarium/data managers and (2) to announce the online presence of an official taxonomy that will define the scope of the newly formed International Committee for the Nomenclature of Lichens and Allied Fungi (ICNLAF). The online version of the taxonomy presented here will continue to evolve along with our understanding of the organisms. Additionally, the subfamily Fissurinoideae Rivas Plata, Lücking and Lumbsch is elevated to the rank of family as Fissurinaceae. KEYWORDS. – higher-level taxonomy, lichen-forming fungi, lichenized fungi, phylogeny INTRODUCTION Traditionally, lichen herbaria have been arranged alphabetically, a scheme that stands in stark contrast to the phylogenetic scheme used by nearly all vascular plant herbaria. The justification typically given for this practice is that lichen taxonomy is too unstable to establish a reasonable system of classification. However, recent leaps forward in our understanding of the higher-level classification of fungi, driven primarily by the NSF-funded Assembling the Fungal Tree of Life (AFToL) project (Lutzoni et al. 2004), have caused the taxonomy of lichen-forming and allied fungi to increase significantly in stability. This is especially true within the class Lecanoromycetes, the main group of lichen-forming fungi (Miadlikowska et al. -

Crittendenia Gen. Nov., a New Lichenicolous Lineage in the Agaricostilbomycetes (Pucciniomycotina), and a Review of the Biology
The Lichenologist (2021), 53, 103–116 doi:10.1017/S002428292000033X Standard Paper Crittendenia gen. nov., a new lichenicolous lineage in the Agaricostilbomycetes (Pucciniomycotina), and a review of the biology, phylogeny and classification of lichenicolous heterobasidiomycetes Ana M. Millanes1, Paul Diederich2, Martin Westberg3 and Mats Wedin4 1Departamento de Biología y Geología, Física y Química Inorgánica, Universidad Rey Juan Carlos, E-28933 Móstoles, Spain; 2Musée national d’histoire naturelle, 25 rue Munster, L-2160 Luxembourg; 3Museum of Evolution, Norbyvägen 16, SE-75236 Uppsala, Sweden and 4Department of Botany, Swedish Museum of Natural History, P.O. Box 50007, SE-10405 Stockholm, Sweden Abstract The lichenicolous ‘heterobasidiomycetes’ belong in the Tremellomycetes (Agaricomycotina) and in the Pucciniomycotina. In this paper, we provide an introduction and review of these lichenicolous taxa, focusing on recent studies and novelties of their classification, phylogeny and evolution. Lichen-inhabiting fungi in the Pucciniomycotina are represented by only a small number of species included in the genera Chionosphaera, Cyphobasidium and Lichenozyma. The phylogenetic position of the lichenicolous representatives of Chionosphaera has, however, never been investigated by molecular methods. Phylogenetic analyses using the nuclear SSU, ITS, and LSU ribosomal DNA mar- kers reveal that the lichenicolous members of Chionosphaera form a monophyletic group in the Pucciniomycotina, distinct from Chionosphaera and outside the Chionosphaeraceae. The new genus Crittendenia is described to accommodate these lichen-inhabiting spe- cies. Crittendenia is characterized by minute synnemata-like basidiomata, the presence of clamp connections and aseptate tubular basidia from which 4–7 spores discharge passively, often in groups. Crittendenia, Cyphobasidium and Lichenozyma are the only lichenicolous lineages known so far in the Pucciniomycotina, whereas Chionosphaera does not include any lichenicolous taxa. -

Epiphytic Seed Microbiomes of Wheat, Canola, and Lentil
EPIPHYTIC SEED MICROBIOMES OF WHEAT, CANOLA, AND LENTIL A Thesis Submitted to the College of Graduate and Postdoctoral Studies In Partial Fulfillment of the Requirements For the Degree of Doctor of Philosophy In the Department of Food and Bioproduct Sciences University of Saskatchewan Saskatoon By Zayda Piedad Morales Moreira © Copyright Zayda Piedad Morales Moreira, June, 2021. All rights reserved. Unless otherwise noted, copyright of the material in this thesis belongs to the author PERMISSION TO USE In presenting this thesis in partial fulfilment of the requirements for a Postgraduate degree from the University of Saskatchewan, I agree that the Libraries of this University may make it freely available for inspection. I further agree that permission for copying of this thesis in any manner, in whole or in part, for scholarly purposes may be granted by the professor or professors who supervised my thesis work or, in their absence, by the Head of the Department or the Dean of the College in which my thesis work was done. It is understood that any copying, publication, or use of this thesis or parts thereof for financial gain shall not be allowed without my written permission. It is also understood that due recognition shall be given to me and to the University of Saskatchewan in any scholarly use which may be made of any material in my thesis. Requests for permission to copy or to make other use of material in this thesis in whole or part should be addressed to: Head of the Department of Food and Bioproduct Sciences University of Saskatchewan 51 Campus Drive University of Saskatchewan Saskatoon, Saskatchewan, S7N 5A8 Canada OR Dean of the College of Graduate and Postdoctoral Studies University of Saskatchewan 107 Administration Place Saskatoon, Saskatchewan S7N 5A2 Canada i ABSTRACT Microorganisms are found colonizing all plant organs including seeds. -

Lichens and Associated Fungi from Glacier Bay National Park, Alaska
The Lichenologist (2020), 52,61–181 doi:10.1017/S0024282920000079 Standard Paper Lichens and associated fungi from Glacier Bay National Park, Alaska Toby Spribille1,2,3 , Alan M. Fryday4 , Sergio Pérez-Ortega5 , Måns Svensson6, Tor Tønsberg7, Stefan Ekman6 , Håkon Holien8,9, Philipp Resl10 , Kevin Schneider11, Edith Stabentheiner2, Holger Thüs12,13 , Jan Vondrák14,15 and Lewis Sharman16 1Department of Biological Sciences, CW405, University of Alberta, Edmonton, Alberta T6G 2R3, Canada; 2Department of Plant Sciences, Institute of Biology, University of Graz, NAWI Graz, Holteigasse 6, 8010 Graz, Austria; 3Division of Biological Sciences, University of Montana, 32 Campus Drive, Missoula, Montana 59812, USA; 4Herbarium, Department of Plant Biology, Michigan State University, East Lansing, Michigan 48824, USA; 5Real Jardín Botánico (CSIC), Departamento de Micología, Calle Claudio Moyano 1, E-28014 Madrid, Spain; 6Museum of Evolution, Uppsala University, Norbyvägen 16, SE-75236 Uppsala, Sweden; 7Department of Natural History, University Museum of Bergen Allégt. 41, P.O. Box 7800, N-5020 Bergen, Norway; 8Faculty of Bioscience and Aquaculture, Nord University, Box 2501, NO-7729 Steinkjer, Norway; 9NTNU University Museum, Norwegian University of Science and Technology, NO-7491 Trondheim, Norway; 10Faculty of Biology, Department I, Systematic Botany and Mycology, University of Munich (LMU), Menzinger Straße 67, 80638 München, Germany; 11Institute of Biodiversity, Animal Health and Comparative Medicine, College of Medical, Veterinary and Life Sciences, University of Glasgow, Glasgow G12 8QQ, UK; 12Botany Department, State Museum of Natural History Stuttgart, Rosenstein 1, 70191 Stuttgart, Germany; 13Natural History Museum, Cromwell Road, London SW7 5BD, UK; 14Institute of Botany of the Czech Academy of Sciences, Zámek 1, 252 43 Průhonice, Czech Republic; 15Department of Botany, Faculty of Science, University of South Bohemia, Branišovská 1760, CZ-370 05 České Budějovice, Czech Republic and 16Glacier Bay National Park & Preserve, P.O. -

The Fungi Constitute a Major Eukary- Members of the Monophyletic Kingdom Fungi ( Fig
American Journal of Botany 98(3): 426–438. 2011. T HE FUNGI: 1, 2, 3 … 5.1 MILLION SPECIES? 1 Meredith Blackwell 2 Department of Biological Sciences; Louisiana State University; Baton Rouge, Louisiana 70803 USA • Premise of the study: Fungi are major decomposers in certain ecosystems and essential associates of many organisms. They provide enzymes and drugs and serve as experimental organisms. In 1991, a landmark paper estimated that there are 1.5 million fungi on the Earth. Because only 70 000 fungi had been described at that time, the estimate has been the impetus to search for previously unknown fungi. Fungal habitats include soil, water, and organisms that may harbor large numbers of understudied fungi, estimated to outnumber plants by at least 6 to 1. More recent estimates based on high-throughput sequencing methods suggest that as many as 5.1 million fungal species exist. • Methods: Technological advances make it possible to apply molecular methods to develop a stable classifi cation and to dis- cover and identify fungal taxa. • Key results: Molecular methods have dramatically increased our knowledge of Fungi in less than 20 years, revealing a mono- phyletic kingdom and increased diversity among early-diverging lineages. Mycologists are making signifi cant advances in species discovery, but many fungi remain to be discovered. • Conclusions: Fungi are essential to the survival of many groups of organisms with which they form associations. They also attract attention as predators of invertebrate animals, pathogens of potatoes and rice and humans and bats, killers of frogs and crayfi sh, producers of secondary metabolites to lower cholesterol, and subjects of prize-winning research. -

The Phylogeny of Plant and Animal Pathogens in the Ascomycota
Physiological and Molecular Plant Pathology (2001) 59, 165±187 doi:10.1006/pmpp.2001.0355, available online at http://www.idealibrary.com on MINI-REVIEW The phylogeny of plant and animal pathogens in the Ascomycota MARY L. BERBEE* Department of Botany, University of British Columbia, 6270 University Blvd, Vancouver, BC V6T 1Z4, Canada (Accepted for publication August 2001) What makes a fungus pathogenic? In this review, phylogenetic inference is used to speculate on the evolution of plant and animal pathogens in the fungal Phylum Ascomycota. A phylogeny is presented using 297 18S ribosomal DNA sequences from GenBank and it is shown that most known plant pathogens are concentrated in four classes in the Ascomycota. Animal pathogens are also concentrated, but in two ascomycete classes that contain few, if any, plant pathogens. Rather than appearing as a constant character of a class, the ability to cause disease in plants and animals was gained and lost repeatedly. The genes that code for some traits involved in pathogenicity or virulence have been cloned and characterized, and so the evolutionary relationships of a few of the genes for enzymes and toxins known to play roles in diseases were explored. In general, these genes are too narrowly distributed and too recent in origin to explain the broad patterns of origin of pathogens. Co-evolution could potentially be part of an explanation for phylogenetic patterns of pathogenesis. Robust phylogenies not only of the fungi, but also of host plants and animals are becoming available, allowing for critical analysis of the nature of co-evolutionary warfare. Host animals, particularly human hosts have had little obvious eect on fungal evolution and most cases of fungal disease in humans appear to represent an evolutionary dead end for the fungus. -

Crittendenia Gen. Nov., a New Lichenicolous Lineage in the Agaricostilbomycetes (Pucciniomycotina), and a Review of the Biology
The Lichenologist (2021), 53, 103–116 doi:10.1017/S002428292000033X Standard Paper Crittendenia gen. nov., a new lichenicolous lineage in the Agaricostilbomycetes (Pucciniomycotina), and a review of the biology, phylogeny and classification of lichenicolous heterobasidiomycetes Ana M. Millanes1, Paul Diederich2, Martin Westberg3 and Mats Wedin4 1Departamento de Biología y Geología, Física y Química Inorgánica, Universidad Rey Juan Carlos, E-28933 Móstoles, Spain; 2Musée national d’histoire naturelle, 25 rue Munster, L-2160 Luxembourg; 3Museum of Evolution, Norbyvägen 16, SE-75236 Uppsala, Sweden and 4Department of Botany, Swedish Museum of Natural History, P.O. Box 50007, SE-10405 Stockholm, Sweden Abstract The lichenicolous ‘heterobasidiomycetes’ belong in the Tremellomycetes (Agaricomycotina) and in the Pucciniomycotina. In this paper, we provide an introduction and review of these lichenicolous taxa, focusing on recent studies and novelties of their classification, phylogeny and evolution. Lichen-inhabiting fungi in the Pucciniomycotina are represented by only a small number of species included in the genera Chionosphaera, Cyphobasidium and Lichenozyma. The phylogenetic position of the lichenicolous representatives of Chionosphaera has, however, never been investigated by molecular methods. Phylogenetic analyses using the nuclear SSU, ITS, and LSU ribosomal DNA mar- kers reveal that the lichenicolous members of Chionosphaera form a monophyletic group in the Pucciniomycotina, distinct from Chionosphaera and outside the Chionosphaeraceae. The new genus Crittendenia is described to accommodate these lichen-inhabiting spe- cies. Crittendenia is characterized by minute synnemata-like basidiomata, the presence of clamp connections and aseptate tubular basidia from which 4–7 spores discharge passively, often in groups. Crittendenia, Cyphobasidium and Lichenozyma are the only lichenicolous lineages known so far in the Pucciniomycotina, whereas Chionosphaera does not include any lichenicolous taxa.