Studies of Genetic Variation at the Kit Locus and White Spotting Patterns in the Horse
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Mcoa Report Final
REPORT TO ROCKY MOUNTAIN HORSE ASSOCIATION GENETIC SURVEY OF THE PMEL17/ SILVER MUTATION WHICH CAUSES MULTIPLE CONGENITAL OCULAR ANOMALIES (MCOA) Final Draft accepted by RMHA Board: 3 October 2020 Prepared by Noah Anderson Chair, Genetics Committee RMHA Assistant Professor of Biology Winona State University Acknowledgements: Our understanding of MCOA results from the efforts of many people. This disorder has been a part of our breed since the beginning. When reading this report, it would be all too easy to forget that the first study happened over 20 years ago with the cooperation of many bold members of the RMHA and the RMHA itself. Rather than letting politics or popularity of their decisions paralyze them, they chose to face MCOA head on. We owe those people a debt of gratitude for doing the right thing by the horse. This study could have stalled out before it even began if it wasn’t for the dogged determination of David Swan and Steve Autry. David and Steve kept us (the genetics committee) corralled until we understood how important understanding MCOA is to the future of our breed. Steve developed the proposal and laid out the experimental design for this project, then carefully shepherded the project until it was near completion. We are fortunate that Steve is continuing to be a guiding light in our genetics committee, as we work to continue the tradition of doing the right thing by the Rocky Mountain Horse. David passed away while this study was being conducted; we will miss his leadership. Mik Fenn kept me swimming in data from our pedigree database of which he is the expert custodian. -

I . the Color Gene C
THE ABC OF COLOR INHERITANCE IN HORSES W. E. CASTLE Division of Genetics, University of California, Berkeley, California Received October, 27, 1947 HE study of color inheritance in horses was begun in the early days of Tgenetics. Indeed many facts concerning it had already been established earlier, by DARWINin his book on “Variation of Animals and Plants under Domestication.” At irregular inteivals since then, new attempts have been made to collect and classify in terms of genetic factors the records contained in stud books concerning the colors of colts in relation to the colors of their sires and dams. A full bibliography is given by CREWand BuCHANAN-SMITH (19301. By such studies, we have acquired very full information as to what color a colt may be expected to have, when the color of its parents and grandparents is known. This knowledge is empirical rather than experimental in nature. For horses being slow breeding and expensive are rarely available for direct experi- mental study, such as can be made with the small laboratory mammals, mice, rats, rabbits and guinea pigs. We have definite information that color inheritance in horses involves the existence of mutant genes similar to those demonstrated by experimental studies to be involved in color inheritance of other mammals. But the horse genes have been given special names, as they were successively discovered, and it is difficult at present to correlate them with the better known names and geneic symbols used by the experimental breeders. The present paper is an attempt to make such a correlation. Just as in morphological studies comparative anatomy was found useful and still is used to establish homologies between systems of organs, so in mammalian genetics, a comparative study of gene action in the production of coat colors and color patterns may also be of value. -

List of Horse Breeds 1 List of Horse Breeds
List of horse breeds 1 List of horse breeds This page is a list of horse and pony breeds, and also includes terms used to describe types of horse that are not breeds but are commonly mistaken for breeds. While there is no scientifically accepted definition of the term "breed,"[1] a breed is defined generally as having distinct true-breeding characteristics over a number of generations; its members may be called "purebred". In most cases, bloodlines of horse breeds are recorded with a breed registry. However, in horses, the concept is somewhat flexible, as open stud books are created for developing horse breeds that are not yet fully true-breeding. Registries also are considered the authority as to whether a given breed is listed as Light or saddle horse breeds a "horse" or a "pony". There are also a number of "color breed", sport horse, and gaited horse registries for horses with various phenotypes or other traits, which admit any animal fitting a given set of physical characteristics, even if there is little or no evidence of the trait being a true-breeding characteristic. Other recording entities or specialty organizations may recognize horses from multiple breeds, thus, for the purposes of this article, such animals are classified as a "type" rather than a "breed". The breeds and types listed here are those that already have a Wikipedia article. For a more extensive list, see the List of all horse breeds in DAD-IS. Heavy or draft horse breeds For additional information, see horse breed, horse breeding and the individual articles listed below. -

Identification of Horses Booklet
colours & marking booklet_10366 21/7/08 14:32 Page 1 1 Introduction The first edition of this booklet was made available to the veterinary profession in 1930. It was in the form of a report from a Sub-Committee, which had been set up by the Council of the RCVS in 1928, to prepare a system of description, colours and markings, etc. of horses for the purpose of identifying individual animals. Since that date there have been several revised editions. Amendments have been made in the light of experience and in response to the changing demands of the equine industry. The significant increase in the international movement of horses and the insistence by a growing number of organisations which hold shows, gymkhanas, events and other competitive functions that horse owners should supply proof of vaccination against equine influenza, have resulted in greater numbers of practitioners being asked to supply the relevant certification. Additionally DEFRA legislation came into force in 2004 requiring all horses to have a passport containing an accurate set of markings. This latest edition (published May 2008) has been produced by Weatherbys, in conjunction with the RCVS and BEVA, in part as a result of the increased use internationally of microchip transponders to identify horses. Nevertheless, the passport, with its recording of a horse’s colour and markings, remains the essential means of identification for Thoroughbred and Non-Thoroughbred horses and ponies under the implementation Regulation of the EC Directives (Commission Regulation (EC) No. 504/2008 of 6 June 2008 and Council Directives 90/426/EEC and 90/427 EEC. -

Háziasítás (Zöldág
2019.09.26. UNIVERSITY OF VETERINARY MEDICINE - BUDAPEST DEPARTMENT OF ANIMAL BREEDING NUTRITION AND LABORATORY ANIMAL SCIENCE SECTION FOR ANIMAL BREEDING AND GENETICS Gene effects, Mendelian exceptions László Zöldág prof. emer. Mendel’s laws in simple inheritance (for monogenic traits!) Preconditions: monogenic, single locus, location on different chromosomes, not X-linked! 1. Homozygosity of parents • 2. Uniformity and reciprocity (criss-cross) in F1 generation: – identity of genotype and phenotype in case of homozygous parents, – independently which of parents carries the dominant or the recessive genes. • 3. Segregation in F2 generation (1:2:1, 3:1): reappearance of parental characteristics in F2 generation • 4. Independent and free segregation and new combinations of genes at diff. loci (9:3:3:1 segregation in case of two loci): inter-chromosomal or Mendelian recombination in mono-, di-, tri- and polyhybrid crossings. 1 2019.09.26. Exceptions for Mendelian Rules: Terminology • Varied expressivity of dominant genes (white spotting and grey colour in horses, epigenetics) • Incomplete penetrance of dominant genes (%, canine JKD) • Multiple alleles, allelic polymorphism (on the same locus) • Epistasis (interlocal interaction of genes, coat colours - oligogenie) – Complementary or epistatic oligogenie (comb types of fowl, coat colours) • Genetic heterogeneity (mimic genes, diff. genotypes the same phenotype) • Linkage and crossing over (intrachromosomale recombination) • Pleiotropy (dosage effect or linkage, „side” effects of genes, Polledness in goats, lethal genes, FecX in sheep) • Sex-X(Z)-linked inheritance (sexing, auto-sexing chicks, X-linked diseases) • Uniparental inheritance (genomic imprinting, maternal inheritance – mtDNA) Changes in gene effects: Mendelian inheritance is not true (?) • Expressivity of genes: – Varied manifestation of dominant genes in heterozygotes, and recessive genes in homozygotes. -

Show Spur Steel Gold Silver Bronze Platinum Diamond Spur Spur
Corporate Partner Program Show Spur Steel Spur Bronze Spur Silver Spur Gold Spur Platinum Spur Diamond Spur the purpose of breeding superior colored horses. The registry they About PtHA created is the basis of what is The Pinto horse originated in the wild herds to develop a stockier now known as the Pinto Horse Spain and was introduced to North and heavier muscled horse that Association of America Inc. America by Spanish and other would be more suited to the rugged It took several years and several European explorers. and arduous conditions. more organizations to form today’s The Spanish explorers brought Often referred to as piebald or PtHA. Tired of the discrimination over Barb horses that had been skewbald horses in literature about Pintos received in the show ring, crossed with other European breeds the Wild West, the Pinto horse Kay Heikens, Helen Hammond and including Russian and Arabian was a favorite among American other Pinto lovers decided to start strains, which are thought to give cowboys and Native Americans. a new organization and registry. the horses their color patterns. Many famous Pintos include The determined group finalized When the Spanish herds were Tonto’s Scout, Little Joe’s Cochise their hard work on May 18, 1956, brought to North America, these and Frank Hopkins’ Hidalgo. in New Jersey when the PtHA was horses mixed with the wild horses The Pinto Horse Association incorporated. and were later domesticated by the started from a grassroots movement Today, the Pinto Family Native Americans. to selectively breed horses for good encompasses more than 47,000 Later, when the West was being color and conformation. -

The Color Breed 2017 Ptha Youth Judging Contest
the Color Breed 2017 PtHA Youth Judging Contest Youth from all over the nation gathered at the Built Ford Tough Livestock Arena for the 4th annual Youth Judging Contest. Participants judged Western Pleasure, Ranch Riding, Western Horsemanship, Western Riding, Hunter Under Saddle, Hunt Seat Equitation, Halter Performance Geldings and Halter Performance Mares classes. Not only did participants judge classes they also gave reasons for selected classes. The Youth Judging Contest, held in conjunction with the Pinto World Championship, consisted of team competition and individual competition. This year there were over 10 teams that competed. photo by: Pinto Horse Association Illinois Quarter horse took the top spot Above: the Illinois Quarter Horse Team in the Senior Youth Overall division and Below: Chouteau Team the Chouteau team became the overall winner within the Junior Youth division. The top teams took home Gist Silversmith belt buckles, trophies and several plaques. The PtHA would like to say a big thank you to all of those who donated their time to exhibit their horse during the Youth Judging Contest. This event would not be possible without members’ support and willingness to volunteer. Congratulations to all 2017 Youth Judging Contest Winners! TODAY’s ClAsses, Tomorrow’s CHAMPIONS 28 OP - Hunter Over Fences In-Hand, Miniature 7:30 a.m. Arena 1 29 OP - Hunter Over Fences In-Hand, B Mini 1 YA Jr - English Pleasure, ST/HN Type Horse 30 OP - Hunter Over Fences In-Hand, Solid Miniature/B Mini 2 YA Sr - English Pleasure, ST/HN Type Horse -

Homozygous Tobiano and Homozygous Black Could Be Winners for Your Breeding Program, If You Know How to Play Your Cards
By IRENE STAMATELAKYS Homozygous tobiano and homozygous black could be winners for your breeding program, if you know how to play your cards. L L I T S K C O T S N N A Y S E T R U O C n poker, a pair is not much to brag gets one of the pair from the sire and the in equine color genetics. If your goal about. Two pairs are just a hair bet - other of the pair from the dam.” is a black foal, and you’ve drawn the ter. But in equine color genetics, a Every gene has an address—a spe - Agouti allele, you’re out of luck. pair—or, even better, two—could cific site on a specific chromosome. be one of the best hands you’ll ever We call this address a locus—plural The Agouti effect hold. We’re talking about a sure bet— being loci. Quite often, geneticists use Approximately 20 percent of horses a pair of tobiano or black genes. the locus name to refer to a gene. registered with the APHA are bay. If Any Paint breeder will tell you that When a gene comes in different you also include the colors derived producing a quality foal that will forms, those variations are called alle - from bay—buckskin, dun, bay roan bring in top dollar is a gamble. In this les. For example, there is a tobiano and perlino—almost one-quarter of business, there are no guarantees. But allele and a non-tobiano allele. Either registered Paints carry and express the what if you could reduce some of the one can occur at the tobiano locus, Agouti allele, symbolized by an upper - risk in your breeding program as well but each chromosome can only carry case A. -
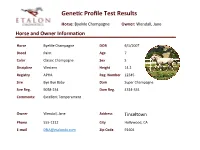
Gene C Profile Test Results
GeneGc Profile Test Results Horse: ByeMe Champagne Owner: Wendall, Jane Horse and Owner Informaon Horse ByeMe Champagne DOB 6/1/2007 Breed Paint Age 7 Color Classic Champagne Sex S Discipline Western Height 14.2 Registry APHA Reg. Number 12345 Sire Bye Bye Baby Dam Super Champagne Sire Reg. 9058-234 Dam Reg. 4314-334 Comments: Excellent Temperament Owner Wendall, Jane Address Tinseltown Phone 555-1212 City Hollywood, CA E-mail [email protected] Zip Code 91604 GeneGc Profile Test Results Horse: ByeMe Champagne Owner: Wendall, Jane Results Summary Coat Color : ByeMe Champagne has one Black allele and one Red allele making the Base coat appear Black. Also detected were single Champagne and Cream alleles; likely resulUng in a rare Champagne Cream color. One copy of the Frame Overo allele is also detected, indicang underlying white patches (hidden By CH). As a result of single gene copies in each of the following, he has a 50% chance of passing Black or Red, Cream and/or Champagne, AgouU and/or Frame Overo alleles to his offspring. Allele Summary: Aa, Ee, Ch, Cr, LWO/n Traits: ByeMe Champagne is a not a carrier of any known recessive disease genes. CauUon is recommended however, as any mare Bred to him should Be Frame Overo negave as to avoid a 25% chance of foal death (+/+ LWO results in a lethal condiUon at Birth). He may also throw Gaited foals when Bred to Gaited (+) mares. Notes: Please note that your analysis is ongoing and may include some regions marked with an asterisk denoUng the following: * Discovery – This gene detecUon is in the -

The Base Colors: Black and Chestnut the Tail, Called “Foal Fringes.”The Lower Legs Can Be So Pale That It Is Let’S Begin with the Base Colors
Foal Color 4.08 3/20/08 2:18 PM Page 44 he safe arrival of a newborn foal is cause for celebration. months the sun bleaches the foal’s birth coat, altering its appear- After checking to make sure all is well with the mare and ance even more. Other environmental issues, such as type and her new addition, the questions start to fly. What gender quality of feed, also can have a profound effect on color. And as we is it? Which traits did the foal get from each parent? And shall see, some colors do change drastically in appearance with Twhat color is it, anyway? Many times this question is not easily age, such as gray and the roany type of sabino. Finally, when the answered unless the breeder has seen many foals, of many colors, foal shed occurs, the new color coming in often looks dramatical- throughout many foaling seasons. In the landmark 1939 movie, ly dark. Is it any wonder that so many foals are registered an incor- “The Wizard of Oz,” MGM used gelatin to dye the “Horse of a rect—and sometimes genetically impossible—color each year? Different Color,” but Mother Nature does a darn good job of cre- So how do you identify your foal’s color? First, let’s keep some ating the same spectacular special effects on her foals! basic rules of genetics in mind. Two chestnuts will only produce The foal’s color from birth to the foal shed (which generally chestnut; horses of the cream, dun, and silver dilutions must have occurs between three and four months of age) can change due to had at least one parent with that particular dilution themselves; many factors, prompting some breeders to describe their foal as and grays must always have one gray parent. -

Genetic Test Results
Genetic Profile Test Results HORSE ID: 041418 003 Horse: Merlin PACK: 1 Owner: Alecia Baxley Horse and Owner Information Horse Merlin DOB 2018-04-04 Breed Paint (Overo) Age 0 years, 0 months Color Chestnut/Overo Sex Stallion Discipline Halter Height . Registry APHA Reg Number Pending Sire Ententions Dam Shes Forever Cool Sire Reg & No. Quarter 5605512 Dam Reg & No. APHA 996357 Comments . Owner Alecia Baxley Address 968 County Road 2117 Phone 281 592 6550 City, State Cleveland, TX Email [email protected] Postal Code 77327 [email protected] 650.380.2995 www.EtalonDx.com Genetic Profile Test Results HORSE ID: 041418 003 Horse: Merlin PACK: 1 Owner: Alecia Baxley Results Summary Coat Color: Merlin has two Red alleles and no Black, indicating his base coat color appears Red. One Dominant White 20 allele and one Frame/Lethal White Overo allele was also detected which may result in White markings. As a result of the allele count in each of the following, he has a minimum 100% chance of passing Red, and 50% Dominant White 20 and/or Frame/Lethal White Overo to any offspring. Allele aa, ee, W20/n, LWO/n, HYPP/n, CC (Sprint Type) Summary: Traits: Merlin's testing indicated the presence of one Frame/Lethal White Overo (LWO) allele resulting in “Carrier” status. Caution is recommended when breeding to avoid another carrier and thus, 25% chance of foal death. His testing also indicated the presence of one Hyperkalemic Periodic Paralysis (HYPP) allele indicating “Carrier” and “Possibly Affected” status. Please consult with your veterinarian regarding any medical questions or advice. -

Culturing of Melanocytes from the Equine Hair Follicle Outer Root Sheath
processes Article Culturing of Melanocytes from the Equine Hair Follicle Outer Root Sheath Hanluo Li 1,† , Jule Kristin Michler 2,† , Alexander Bartella 1 , Anna Katharina Sander 1, Sebastian Gaus 1, Sebastian Hahnel 3, Rüdiger Zimmerer 1, Jan-Christoph Simon 4, Vuk Savkovic 1,*,‡ and Bernd Lethaus 1,‡ 1 Department of Cranial Maxillofacial Plastic Surgery, University Hospital Leipzig, 04103 Leipzig, Germany; [email protected] (H.L.); [email protected] (A.B.); [email protected] (A.K.S.); [email protected] (S.G.); [email protected] (R.Z.); [email protected] (B.L.) 2 Institute of Veterinary Anatomy, University of Leipzig, 04103 Leipzig, Germany; [email protected] 3 Polyclinic for Dental Prosthetics and Material Sciences, University Hospital Leipzig, 04103 Leipzig, Germany; [email protected] 4 Clinic for Dermatology, Venereology and Allergology, University Hospital Leipzig, 04103 Leipzig, Germany; [email protected] * Correspondence: [email protected]; Tel.: +49-341-97-21115 † The first two authors contributed equally to this work. ‡ These authors contributed equally to this work. Abstract: Hair follicles harbor a heterogeneous regenerative cell pool and represent a putative low- to-non-invasively available source of stem cells. We previously reported a technology for culturing human melanocytes from the hair follicle outer root sheath (ORS) for autologous pigmentation of tissue engineered skin equivalents. This study translated the ORS technology to horses. We de-veloped a culture of equine melanocytes from the ORS (eMORS) from equine forelock hair follicles cultured by means of an analogue human hair follicle-based in vitro methodology.