N-Succinylamino Acid Racemases
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Transcriptomic Characterization of Bradyrhizobium Diazoefficiens
International Journal of Molecular Sciences Article Transcriptomic Characterization of Bradyrhizobium diazoefficiens Bacteroids Reveals a Post-Symbiotic, Hemibiotrophic-Like Lifestyle of the Bacteria within Senescing Soybean Nodules Sooyoung Franck 1, Kent N. Strodtman 1 , Jing Qiu 2 and David W. Emerich 1,* 1 Division of Biochemistry, University of Missouri, Columbia, MO 65211, USA; [email protected] (S.F.); [email protected] (K.N.S.) 2 Applied Economics and Statistics, University of Delaware, Newark, DE 19716, USA; [email protected] * Correspondence: [email protected]; Tel: +1-573-882-4252 Received: 8 October 2018; Accepted: 28 November 2018; Published: 7 December 2018 Abstract: The transcriptional activity of Bradyrhizobium diazoefficens isolated from soybean nodules was monitored over the period from symbiosis to late plant nodule senescence. The bacteria retained a near constant level of RNA throughout this period, and the variation in genes demonstrating increased, decreased, and/or patterned transcriptional activity indicates that the bacteria are responding to the changing environment within the nodule as the plant cells progress from an organized cellular structure to an unorganized state of internal decay. The transcriptional variation and persistence of the bacteria suggest that the bacteria are adapting to their environment and acting similar to hemibiotrophs, which survive both as saprophytes on live plant tissues and then as necrophytes on decaying plant tissues. The host plant restrictions of symbiosis make B. diazoefficiens a highly specialized, restricted hemibiotroph. Keywords: bradyrhizobium diazoefficiens; soybean; Glycine max; nitrogen fixation; senescence; transcriptomics; hemibiotroph 1. Introduction Soybean nodules are symbiotic organs that are formed on roots by the complex interaction between soybean plants and rhizobia, nitrogen-fixing bacteria, under nitrogen-limiting conditions. -

Colletotrichum Graminicola</Em>
University of Kentucky UKnowledge Plant Pathology Faculty Publications Plant Pathology 3-8-2016 A Colletotrichum graminicola Mutant Deficient in the Establishment of Biotrophy Reveals Early Transcriptional Events in the Maize Anthracnose Disease Interaction Maria F. Torres University of Kentucky, [email protected] Noushin Ghaffari Texas A&M University Ester A. S. Buiate University of Kentucky, [email protected] Neil Moore University of Kentucky, [email protected] Scott chS wartz Texas A&M University FSeoe nelloxtw pa thige fors aaddndition addal aitutionhorsal works at: https://uknowledge.uky.edu/plantpath_facpub Part of the Bioinformatics Commons, Genomics Commons, Integrative Biology Commons, and Right click to open a feedback form in a new tab to let us know how this document benefits oy u. the Plant Pathology Commons Repository Citation Torres, Maria F.; Ghaffari, Noushin; Buiate, Ester A. S.; Moore, Neil; Schwartz, Scott; Johnson, Charles D.; and Vaillancourt, Lisa J., "A Colletotrichum graminicola Mutant Deficient in the Establishment of Biotrophy Reveals Early Transcriptional Events in the Maize Anthracnose Disease Interaction" (2016). Plant Pathology Faculty Publications. 53. https://uknowledge.uky.edu/plantpath_facpub/53 This Article is brought to you for free and open access by the Plant Pathology at UKnowledge. It has been accepted for inclusion in Plant Pathology Faculty Publications by an authorized administrator of UKnowledge. For more information, please contact [email protected]. Authors Maria F. Torres, Noushin Ghaffari, Ester A. S. Buiate, Neil Moore, Scott chS wartz, Charles D. Johnson, and Lisa J. Vaillancourt A Colletotrichum graminicola Mutant Deficient in the Establishment of Biotrophy Reveals Early Transcriptional Events in the Maize Anthracnose Disease Interaction Notes/Citation Information Published in BMC Genomics, v.17, 202, p. -

Exploring the Chemistry and Evolution of the Isomerases
Exploring the chemistry and evolution of the isomerases Sergio Martínez Cuestaa, Syed Asad Rahmana, and Janet M. Thorntona,1 aEuropean Molecular Biology Laboratory, European Bioinformatics Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge CB10 1SD, United Kingdom Edited by Gregory A. Petsko, Weill Cornell Medical College, New York, NY, and approved January 12, 2016 (received for review May 14, 2015) Isomerization reactions are fundamental in biology, and isomers identifier serves as a bridge between biochemical data and ge- usually differ in their biological role and pharmacological effects. nomic sequences allowing the assignment of enzymatic activity to In this study, we have cataloged the isomerization reactions known genes and proteins in the functional annotation of genomes. to occur in biology using a combination of manual and computa- Isomerases represent one of the six EC classes and are subdivided tional approaches. This method provides a robust basis for compar- into six subclasses, 17 sub-subclasses, and 245 EC numbers cor- A ison and clustering of the reactions into classes. Comparing our responding to around 300 biochemical reactions (Fig. 1 ). results with the Enzyme Commission (EC) classification, the standard Although the catalytic mechanisms of isomerases have already approach to represent enzyme function on the basis of the overall been partially investigated (3, 12, 13), with the flood of new data, an integrated overview of the chemistry of isomerization in bi- chemistry of the catalyzed reaction, expands our understanding of ology is timely. This study combines manual examination of the the biochemistry of isomerization. The grouping of reactions in- chemistry and structures of isomerases with recent developments volving stereoisomerism is straightforward with two distinct types cis-trans in the automatic search and comparison of reactions. -

University of London Thesis
REFERENCE ONLY UNIVERSITY OF LONDON THESIS Degree Year^^0^ Name of Author C O P Y R IG H T This is a thesis accepted for a Higher Degree of the University of London. It is an unpublished typescript and the copyright is held by the author. All persons consulting the thesis must read and abide by the Copyright Declaration below. COPYRIGHT DECLARATION I recognise that the copyright of the above-described thesis rests with the author and that no quotation from it or information derived from it may be published without the prior written consent of the author. LOANS Theses may not be lent to individuals, but the Senate House Library may lend a copy to approved libraries within the United Kingdom, for consultation solely on the premises of those libraries. Application should be made to: Inter-Library Loans, Senate House Library, Senate House, Malet Street, London WC1E 7HU. REPRODUCTION University of London theses may not be reproduced without explicit written permission from the Senate House Library. Enquiries should be addressed to the Theses Section of the Library. Regulations concerning reproduction vary according to the date of acceptance of the thesis and are listed below as guidelines. A. Before 1962. Permission granted only upon the prior written consent of the author. (The Senate House Library will provide addresses where possible). B. 1962- 1974. In many cases the author has agreed to permit copying upon completion of a Copyright Declaration. C. 1975 - 1988. Most theses may be copied upon completion of a Copyright Declaration. D. 1989 onwards. Most theses may be copied. -

257 Absolute Configuration 39, 75 – of L-(+)-Alanine 83
257 Index a L-aminoacylase 246 absolute configuration 39, 75 amount of substance 12 –ofL-(+)-alanine 83 asymmetric atom 39 – by anomalous dispersion effect in X-ray asymmetric disequilibrating transformation crystallography 78 151 – by chemical correlation 79 asymmetric induction 143 – correlation strategies 80 – of second kind 151 – by direct methods 78 asymmetric synthesis 139 – extended sense of 77 asymmetric transformation –ofD-glyceraldehyde 39 – of second kind 151 –ofD-(+)-glyceraldehyde 77 asymmetric transformation of the second kind – by indirect methods 79 173 –ofD-(–)-lactic acid 81 atropisomer 44, 73 – methods of determination 78 atropisomerism 24, 71 – of natural glucose 78, 82 autocatalysis – by predictive calculation of chiroptical data – a special case of organocatalysis 234 79 autocatalytic effect –of(R,R)-(+)-tartaric acid 78, 82 – of a zinc complex 238 achiral bidentate reagent Avogadro constant 12 – purification of enantiomers with 149 axial chirality 39, 44 achiral stationary phase 95 achirality, achiral 24, 29 b aci form 113 Baeyer–Villiger oxidation acylase I 161 – enanatiotope selective 253 AD-mix- 229 – enzymatic 253 AD-mix- 229 – microbial 166 alcohol dehydrogenases – regio- and enanatiomer selective 253 – Prelog’s rule 252 baker’s yeast aldol reactions – regio-, enantiotope and diastereotope – catalyzed with chiral ionic liquid 238 selective reduction with 186 – catalytic, double enantiotope selective 238 bidentate achiral reagent 90 – completely syn diastereoselective 238 Bijvoet, J. M. 78 – stereoselctive 209 BINAL-H 212 allene 44 BINAP amide-imido acid tautomerism 112 – ruthenium complex of 214 amino acids biocatalysis 244 enantiopure, D-andL- 250 – advantages of 245 D-amino acid oxidase 100 – disadvantages of 245 Stereochemistry and Stereoselective Synthesis: An Introduction, First Edition. -

SI Appendix Index 1
SI Appendix Index Calculating chemical attributes using EC-BLAST ................................................................................ 2 Chemical attributes in isomerase reactions ............................................................................................ 3 Bond changes …..................................................................................................................................... 3 Reaction centres …................................................................................................................................. 5 Substrates and products …..................................................................................................................... 6 Comparative analysis …........................................................................................................................ 7 Racemases and epimerases (EC 5.1) ….................................................................................................. 7 Intramolecular oxidoreductases (EC 5.3) …........................................................................................... 8 Intramolecular transferases (EC 5.4) ….................................................................................................. 9 Supporting references …....................................................................................................................... 10 Fig. S1. Overview …............................................................................................................................ -

This Thesis Has Been Submitted in Fulfilment of the Requirements for a Postgraduate Degree (E.G
This thesis has been submitted in fulfilment of the requirements for a postgraduate degree (e.g. PhD, MPhil, DClinPsychol) at the University of Edinburgh. Please note the following terms and conditions of use: This work is protected by copyright and other intellectual property rights, which are retained by the thesis author, unless otherwise stated. A copy can be downloaded for personal non-commercial research or study, without prior permission or charge. This thesis cannot be reproduced or quoted extensively from without first obtaining permission in writing from the author. The content must not be changed in any way or sold commercially in any format or medium without the formal permission of the author. When referring to this work, full bibliographic details including the author, title, awarding institution and date of the thesis must be given. Biocatalytic application of rare PLP- dependent aminotransferases for the synthesis of high value amino acids and amines Annabel Serpico A Thesis Submitted for the Degree of Doctor of Philosophy The University of Edinburgh 2018 Lay Summary Traditional chemical approaches for the production of compounds often rely on harsh conditions such as high temperature and pressure and create toxic waste and pollutants. Moreover, these reactions are often non efficient, creating unwanted side products that need to be disposed of. A very important branch of chemistry, called green chemistry, supports the design of products and processes that use the least amount of dangerous substances as they can. An important aspect of green chemistry is biocatalysis, which consists of the use of biological systems, either whole cells or enzymes for the production of compounds. -

Natural Occurrence and Industrial Applications of D-Amino Acids: an Overview
CHEMISTRY & BIODIVERSITY – Vol. 7 (2010) 1531 REVIEW Natural Occurrence and Industrial Applications of d-Amino Acids: An Overview by Sergio Martnez-Rodrguez*, Ana Isabel Martnez-Go´ mez, Felipe Rodrguez-Vico, Josefa Mara Clemente-Jime´nez, Francisco Javier Las Heras-Va´zquez* Dpto. Qumica Fsica, Bioqumica y Qumica Inorga´nica, Universidad de Almera, Edificio CITE I, Carretera de Sacramento s/n, 04120 La Can˜ ada de San Urbano, Almera, Spain (S. M.-R.: phone: þ34950015850; fax: þ34950015615; F. J. H.-V.: phone: þ34950015055; fax: þ34950015615) Interest in d-amino acids has increased in recent decades with the development of new analytical methods highlighting their presence in all kingdoms of life. Their involvement in physiological functions, and the presence of metabolic routes for their synthesis and degradation have been shown. Furthermore, d-amino acids are gaining considerable importance in the pharmaceutical industry. The immense amount of information scattered throughout the literature makes it difficult to achieve a general overview of their applications. This review summarizes the state-of-the-art on d-amino acid applications and occurrence, providing both established and neophyte researchers with a comprehensive introduction to this topic. Introduction. – Since Miller established in 1953 that amino acids were most probably present in the primitive Earths atmosphere [1], one of the largest mysteries in science has been why evolution chose the l-isomer of a-amino acids for the emergence of life. While, from the chemical-physical point of view, enantiomeric enhancement in the prebiotic scenario of l-amino acids is plausible [2][3], the most broadly accepted theory is that primitive life acquired polymers by an as yet unknown mechanism. -

Efficient Production of Enantiopure D-Lysine from L-Lysine by a Two
Article Efficient Production of Enantiopure D‐Lysine from L‐Lysine by a Two‐Enzyme Cascade System Xin Wang, Li Yang, Weijia Cao, Hanxiao Ying, Kequan Chen * and Pingkai Ouyang State Key Laboratory of Materials‐Oriented Chemical Engineering, College of Biotechnology and Pharmaceutical Engineering, Nanjing Tech University, Nanjing 211816, Jiangsu, China; [email protected] (X.W.); [email protected] (L.Y.); [email protected] (W.C.); [email protected] (H.Y.); [email protected] (P.O.) * Correspondence: [email protected]; Tel.: +86‐25‐5813‐9386 Academic Editors: Jose M. Palomo and Cesar Mateo Received: 24 September 2016; Accepted: 25 October 2016; Published: 30 October 2016 Abstract: The microbial production of D‐lysine has been of great interest as a medicinal raw material. Here, a two‐step process for D‐lysine production from L‐lysine by the successive microbial racemization and asymmetric degradation with lysine racemase and decarboxylase was developed. The whole‐cell activities of engineered Escherichia coli expressing racemases from the strains Proteus mirabilis (LYR) and Lactobacillus paracasei (AAR) were first investigated comparatively. When the strain BL21‐LYR with higher racemization activity was employed, L‐lysine was rapidly racemized to give DL‐lysine, and the D‐lysine yield was approximately 48% after 0.5 h. Next, L‐lysine was selectively catabolized to generate cadaverine by lysine decarboxylase. The comparative analysis of the decarboxylation activities of resting whole cells, permeabilized cells, and crude enzyme revealed that the crude enzyme was the best biocatalyst for enantiopure D‐lysine production. The reaction temperature, pH, metal ion additive, and pyridoxal 5′‐phosphate content of this two‐step production process were subsequently optimized. -

Of Pseudomonas Sp. Strain NS671 Expressed In
JOURNAL OF BACTERIOLOGY, Dec. 1992, p. 7989-7995 Vol. 174, No. 24 0021-9193/92/247989-07$02.00/0 Copyright X) 1992, American Society for Microbiology Purification and Characterization of the Hydantoin Racemase of Pseudomonas sp. Strain NS671 Expressed in Escherichia coli KEN WATABE,* TAKAHIRO ISHIKAWA, YUKUO MUKOHARA, AND HIROAKI NAKAMURA Odawara Research Center, Nippon Soda Co., Ltd., 345 Takada, Odawara, Kanagawa 250-02, Japan Received 16 July 1992/Accepted 19 October 1992 The hydantoin racemase gene ofPseudomonas sp. strain NS671 had been cloned and expressed in Escherichia coli. Hydantoin racemase was purified from the cell extract of the E. coli strain by phenyl-Sepharose, DEAE-Sephacel, and Sephadex G-200 chromatographies. The purified enzyme had an apparent molecular mass of 32 kDa as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. By gel filtration, a molecular mass of about 190 kDa was found, suggesting that the native enzyme is a hexamer. The optimal conditions for hydantoin racemase activity were pH 9.5 and a temperature of 45°C. The enzyme activity was slightly stimulated by the addition of not only Mn2+ or Co2' but also metal-chelating agents, indicating that the enzyme is not a metalloenzyme. On the other hand, Cu2' and Zn2+ strongly inhibited the enzyme activity. Kinetic studies showed substrate inhibition, and the V.. values for D- and L-5-(2-methylthioethyl)hydantoin were 35.2 and 79.0 ,umol/min/mg of protein, respectively. The purified enzyme did not racemize 5-isopropyl- hydantoin, whereas the cells ofE. coli expressing the enzyme are capable of racemizing it. -
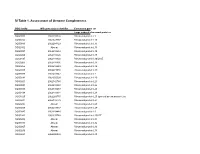
SI Table 1. Assessment of Genome Completeness
SI Table 1. Assessment of Genome Completeness COG family IMG gene object identifier Conserved gene set Large subunit ribosomal proteins COG0081 2062288324 Ribosomal protein L1 COG0244 2062347387 Ribosomal protein L10 COG0080 2062288323 Ribosomal protein L11 COG0102 Absent Ribosomal protein L13 COG0093 2062418832 Ribosomal protein L14 COG0200 2062418826 Ribosomal protein L15 COG0197 2062418838 Ribosomal protein L16/L10E COG0203 2062418836 Ribosomal protein L17 COG0256 2062418829 Ribosomal protein L18 COG0335 2062273558 Ribosomal protein L19 COG0090 2062418842 Ribosomal protein L2 COG0292 2062350539 Ribosomal protein L20 COG0261 2062142780 Ribosomal protein L21 COG0091 2062418840 Ribosomal protein L22 COG0089 2062138283 Ribosomal protein L23 COG0198 2062418834 Ribosomal protein L24 COG1825 2062269715 Ribosomal protein L25 (general stress protein Ctc) COG0211 2062142779 Ribosomal protein L27 COG0227 Absent Ribosomal protein L28 COG0255 2062418837 Ribosomal protein L29 COG0087 2062154483 Ribosomal protein L3 COG1841 2062335748 Ribosomal protein L30/L7E COG0254 Absent Ribosomal protein L31 COG0333 Absent Ribosomal protein L32 COG0267 Absent Ribosomal protein L33 COG0230 Absent Ribosomal protein L34 COG0291 2062350538 Ribosomal protein L35 COG0257 Absent Ribosomal protein L36 COG0088 2062138282 Ribosomal protein L4 COG0094 2062418833 Ribosomal protein L5 COG0097 2062418830 Ribosomal protein L6P/L9E COG0222 2062288326 Ribosomal protein L7/L12 COG0359 2062209880 Ribosomal protein L9 Small subunit ribosomal proteins COG0539 Absent Ribosomal protein -

Assay of the Prostate Cancer Biomarker A-Methylacyl Coenzyme a Racemase (AMACR)
Assay of the Prostate Cancer Biomarker a-Methylacyl Coenzyme A Racemase (AMACR) by Dahmane Ouazia Submitted in partial fulfillment of the requirements for the degree of Master of Science at Dalhousie University Halifax, Nova Scotia June 2008 © Copyright by Dahmane Ouazia, 2008 Library and Bibliotheque et 1*1 Archives Canada Archives Canada Published Heritage Direction du Branch Patrimoine de I'edition 395 Wellington Street 395, rue Wellington Ottawa ON K1A0N4 Ottawa ON K1A0N4 Canada Canada Your file Votre reference ISBN: 978-0-494-43994-4 Our file Notre reference ISBN: 978-0-494-43994-4 NOTICE: AVIS: The author has granted a non L'auteur a accorde une licence non exclusive exclusive license allowing Library permettant a la Bibliotheque et Archives and Archives Canada to reproduce, Canada de reproduire, publier, archiver, publish, archive, preserve, conserve, sauvegarder, conserver, transmettre au public communicate to the public by par telecommunication ou par Plntemet, prefer, telecommunication or on the Internet, distribuer et vendre des theses partout dans loan, distribute and sell theses le monde, a des fins commerciales ou autres, worldwide, for commercial or non sur support microforme, papier, electronique commercial purposes, in microform, et/ou autres formats. paper, electronic and/or any other formats. The author retains copyright L'auteur conserve la propriete du droit d'auteur ownership and moral rights in et des droits moraux qui protege cette these. this thesis. Neither the thesis Ni la these ni des extraits substantiels de nor substantial extracts from it celle-ci ne doivent etre imprimes ou autrement may be printed or otherwise reproduits sans son autorisation.