Differentiation Involved in Early Th1 and Th2 Cell Genome-Wide
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Targeting Hedgehog Signalling Through the Ubiquitylation Process: the Multiple Roles of the HECT-E3 Ligase Itch
cells Review Targeting Hedgehog Signalling through the Ubiquitylation Process: The Multiple Roles of the HECT-E3 Ligase Itch Paola Infante 1,†, Ludovica Lospinoso Severini 2,† , Flavia Bernardi 2, Francesca Bufalieri 2 and Lucia Di Marcotullio 2,3,* 1 Center for Life NanoScience@Sapienza, Istituto Italiano di Tecnologia, 00161 Rome, Italy; [email protected] 2 Department of Molecular Medicine, University of Rome La Sapienza, 00161 Rome, Italy; [email protected] (L.L.S.); fl[email protected] (F.B.); [email protected] (F.B.) 3 Istituto Pasteur-Fondazione Cenci Bolognetti, University of Rome La Sapienza, 00161 Rome, Italy * Correspondence: [email protected]; Tel.: +39-06-49255657 † These authors contributed equally to this work. Received: 28 December 2018; Accepted: 26 January 2019; Published: 29 January 2019 Abstract: Hedgehog signalling (Hh) is a developmental conserved pathway strongly involved in cancers when deregulated. This important pathway is orchestrated by numerous regulators, transduces through distinct routes and is finely tuned at multiple levels. In this regard, ubiquitylation processes stand as essential for controlling Hh pathway output. Although this post-translational modification governs proteins turnover, it is also implicated in non-proteolytic events, thereby regulating the most important cellular functions. The HECT E3 ligase Itch, well known to control immune response, is emerging to have a pivotal role in tumorigenesis. By illustrating Itch specificities on Hh signalling key components, here we review the role of this HECT E3 ubiquitin ligase in suppressing Hh-dependent tumours and explore its potential as promising target for innovative therapeutic approaches. Keywords: Hedgehog; cancer; ubiquitylation; Itch; Numb; β-arrestin2; GLI1; SuFu; Patched1 1. -

An Immunoevasive Strategy Through Clinically-Relevant Pan-Cancer Genomic and Transcriptomic Alterations of JAK-STAT Signaling Components
bioRxiv preprint doi: https://doi.org/10.1101/576645; this version posted March 14, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. An immunoevasive strategy through clinically-relevant pan-cancer genomic and transcriptomic alterations of JAK-STAT signaling components Wai Hoong Chang1 and Alvina G. Lai1, 1Nuffield Department of Medicine, University of Oxford, Old Road Campus, Oxford, OX3 7FZ, United Kingdom Since its discovery almost three decades ago, the Janus ki- Although cytokines are responsible for inflammation in nase (JAK)-signal transducer and activator of transcription cancer, spontaneous eradication of tumors by endoge- (STAT) pathway has paved the road for understanding inflam- nous immune processes rarely occurs. Moreover, the matory and immunity processes related to a wide range of hu- dynamic interaction between tumor cells and host immu- man pathologies including cancer. Several studies have demon- nity shields tumors from immunological ablation, which strated the importance of JAK-STAT pathway components in overall limits the efficacy of immunotherapy in the clinic. regulating tumor initiation and metastatic progression, yet, the extent of how genetic alterations influence patient outcome is far from being understood. Focusing on 133 genes involved in Cytokines can be pro- or anti-inflammatory and are inter- JAK-STAT signaling, we found that copy number alterations dependent on each other’s function to maintain immune underpin transcriptional dysregulation that differs within and homeostasis(3). -

S41467-020-18249-3.Pdf
ARTICLE https://doi.org/10.1038/s41467-020-18249-3 OPEN Pharmacologically reversible zonation-dependent endothelial cell transcriptomic changes with neurodegenerative disease associations in the aged brain Lei Zhao1,2,17, Zhongqi Li 1,2,17, Joaquim S. L. Vong2,3,17, Xinyi Chen1,2, Hei-Ming Lai1,2,4,5,6, Leo Y. C. Yan1,2, Junzhe Huang1,2, Samuel K. H. Sy1,2,7, Xiaoyu Tian 8, Yu Huang 8, Ho Yin Edwin Chan5,9, Hon-Cheong So6,8, ✉ ✉ Wai-Lung Ng 10, Yamei Tang11, Wei-Jye Lin12,13, Vincent C. T. Mok1,5,6,14,15 &HoKo 1,2,4,5,6,8,14,16 1234567890():,; The molecular signatures of cells in the brain have been revealed in unprecedented detail, yet the ageing-associated genome-wide expression changes that may contribute to neurovas- cular dysfunction in neurodegenerative diseases remain elusive. Here, we report zonation- dependent transcriptomic changes in aged mouse brain endothelial cells (ECs), which pro- minently implicate altered immune/cytokine signaling in ECs of all vascular segments, and functional changes impacting the blood–brain barrier (BBB) and glucose/energy metabolism especially in capillary ECs (capECs). An overrepresentation of Alzheimer disease (AD) GWAS genes is evident among the human orthologs of the differentially expressed genes of aged capECs, while comparative analysis revealed a subset of concordantly downregulated, functionally important genes in human AD brains. Treatment with exenatide, a glucagon-like peptide-1 receptor agonist, strongly reverses aged mouse brain EC transcriptomic changes and BBB leakage, with associated attenuation of microglial priming. We thus revealed tran- scriptomic alterations underlying brain EC ageing that are complex yet pharmacologically reversible. -

Impact of Chromosome 9 Numerical Imbalances in Oral Squamous Cell Carcinoma: a Pilot Grid-Based Centromere Analysis
diagnostics Communication Impact of Chromosome 9 Numerical Imbalances in Oral Squamous Cell Carcinoma: A Pilot Grid-Based Centromere Analysis 1, 1, 2, Efthymios Kyrodimos y , Aristeidis Chrysovergis y , Nicholas Mastronikolis y, Evangelos Tsiambas 3,*, Christos Riziotis 4,5,* , Dimitrios Roukas 6, Panagiotis Fotiades 7, Chara Stavraka 8 , Vasileios Ragos 9, Minas Paschopoulos 10 and Vasileios Papanikolaou 1 1 1st ENT Department, Hippocration General Hospital, University of Athens, 115 27 Athens, Greece; [email protected] (E.K.); [email protected] (A.C.); [email protected] (V.P.) 2 ENT Department, Medical School, University of Patras, 265 04 Patras, Greece; [email protected] 3 Department of Cytopathology, 417 Veterans Army Hospital (NIMTS), 115 21 Athens, Greece 4 Theoretical and Physical Chemistry Institute, Photonics for Nanoapplications Laboratory, National Hellenic Research Foundation, 11635 Athens, Greece 5 Defence & Security Research Institute, University of Nicosia, CY-2417 Nicosia, Cyprus 6 Department of Psychiatry, 417 Veterans Army Hospital (NIMTS), 115 21 Athens, Greece; [email protected] 7 Department of Surgery, 424 General Army Hospital, 564 29 Thessaloniki, Greece; [email protected] 8 Department of Medical Oncology, Guy’s and St Thomas National Health System Foundation Trust, London SE1 9RT, UK; [email protected] 9 Department of Maxillofacial Surgery, Medical School, University of Ioannina, 455 00 Ioannina, Greece; [email protected] 10 Department of Obstetrics and Gynaecology, School of Health Sciences, University of Ioannina, 455 00 Ioannina, Greece; [email protected] * Correspondence: [email protected] (E.T.); [email protected] (C.R.); Tel.: +00306946939414 (E.T.) These authors are equally contributed. y Received: 16 June 2020; Accepted: 14 July 2020; Published: 21 July 2020 Abstract: Oral squamous cell carcinoma (OSCC) is considered an aggressive malignancy, mainly due to its increased propensity to provide local and distant lymph node metastases. -

Cutting Edge: Expression of IRF8 in Gastric Epithelial Cells Confers
Cutting Edge: Expression of IRF8 in Gastric Epithelial Cells Confers Protective Innate Immunity against Helicobacter pylori Infection This information is current as of September 27, 2021. Ming Yan, Hongsheng Wang, Jiafang Sun, Wei Liao, Peng Li, Yin Zhu, Chengfu Xu, Jungsoo Joo, Yan Sun, Sadia Abbasi, Alexander Kovalchuk, Nonghua Lv, Warren J. Leonard and Herbert C. Morse III J Immunol published online 3 February 2016 Downloaded from http://www.jimmunol.org/content/early/2016/02/02/jimmun ol.1500766 http://www.jimmunol.org/ Supplementary http://www.jimmunol.org/content/suppl/2016/02/02/jimmunol.150076 Material 6.DCSupplemental Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists by guest on September 27, 2021 • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2016 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. Published February 3, 2016, doi:10.4049/jimmunol.1500766 Th eJournal of Cutting Edge Immunology Cutting Edge: Expression of IRF8 in Gastric Epithelial Cells Confers Protective Innate Immunity against Helicobacter pylori Infection x x Ming Yan,*,1 Hongsheng Wang,†,1 Jiafang Sun,† Wei Liao,‡, Peng Li,‡, { † Yin Zhu, ChengfuXu,*JungsooJoo,*YanSun,*SadiaAbbasi,{ x Alexander Kovalchuk,† Nonghua Lv, Warren J. -

Table S1| Differential Expression Analysis of the Atopy Transcriptome
Table S1| Differential expression analysis of the atopy transcriptome in CD4+ T-cell responses to allergens in atopic and nonatopic subjects Probe ID S.test Gene Symbol Gene Description Chromosome Statistic Location 7994280 10.32 IL4R Interleukin 4 receptor 16p11.2-12.1 8143383 8.95 --- --- --- 7974689 8.50 DACT1 Dapper, antagonist of beta-catenin, homolog 1 14q23.1 8102415 7.59 CAMK2D Calcium/calmodulin-dependent protein kinase II delta 4q26 7950743 7.58 RAB30 RAB30, member RAS oncogene family 11q12-q14 8136580 7.54 RAB19B GTP-binding protein RAB19B 7q34 8043504 7.45 MAL Mal, T-cell differentiation protein 2cen-q13 8087739 7.27 CISH Cytokine inducible SH2-containing protein 3p21.3 8000413 7.17 NSMCE1 Non-SMC element 1 homolog (S. cerevisiae) 16p12.1 8021301 7.15 RAB27B RAB27B, member RAS oncogene family 18q21.2 8143367 6.83 SLC37A3 Solute carrier family 37 member 3 7q34 8152976 6.65 TMEM71 Transmembrane protein 71 8q24.22 7931914 6.56 IL2R Interleukin 2 receptor, alpha 10p15-p14 8014768 6.43 PLXDC1 Plexin domain containing 1 17q21.1 8056222 6.43 DPP4 Dipeptidyl-peptidase 4 (CD26) 2q24.3 7917697 6.40 GFI1 Growth factor independent 1 1p22 7903507 6.39 FAM102B Family with sequence similarity 102, member B 1p13.3 7968236 5.96 RASL11A RAS-like, family 11, member A --- 7912537 5.95 DHRS3 Dehydrogenase/reductase (SDR family) member 3 1p36.1 7963491 5.83 KRT1 Keratin 1 (epidermolytic hyperkeratosis) 12q12-q13 7903786 5.72 CSF1 Colony stimulating factor 1 (macrophage) 1p21-p13 8019061 5.67 SGSH N-sulfoglucosamine sulfohydrolase (sulfamidase) 17q25.3 -

Molecular Characterization of Acquired Tolerance of Tumor Cells to Picropodophyllin (PPP)
Molecular Characterization of Acquired Tolerance of Tumor Cells to Picropodophyllin (PPP) Jamileh Hashemi1*, Claire Worrall2, Daiana Vasilcanu2,Ma˚rten Frykna¨s3, Luqman Sulaiman1, Mohsen Karimi1, Wen-Hui Weng1¤a, Weng-Onn Lui1, Christina Rudduck1¤b, Magnus Axelson1, Helena Jernberg-Wiklund3, Leonard Girnita2, Olle Larsson2, Catharina Larsson1 1 Department of Molecular Medicine and Surgery, Center for Molecular Medicine, Karolinska Institutet, Karolinska University Hospital, CMM L8:01, Stockholm, Sweden, 2 Department of Oncology-Pathology, Karolinska Institutet, Karolinska University Hospital, CCK R8:04, Stockholm, Sweden, 3 Department of Genetics and Pathology, Rudbeck Laboratory, Uppsala University, Uppsala, Sweden Abstract Background: Picropodophyllin (PPP) is a promising novel anti-neoplastic agent that efficiently kills tumor cells in vitro and causes tumor regression and increased survival in vivo. We have previously reported that PPP treatment induced moderate tolerance in two out of 10 cell lines only, and here report the acquired genomic and expression alterations associated with PPP selection over 1.5 years of treatment. Methodology/Principal Findings: Copy number alterations monitored using metaphase and array-based comparative genomic hybridization analyses revealed largely overlapping alterations in parental and maximally tolerant cells. Gain/ amplification of the MYC and PVT1 loci in 8q24.21 were verified on the chromosome level. Abnormalities observed in connection to PPP treatment included regular gains and losses, as well as homozygous losses in 10q24.1-q24.2 and 12p12.3- p13.2 in one of the lines and amplification at 5q11.2 in the other. Abnormalities observed in both tolerant derivatives include amplification/gain of 5q11.2, gain of 11q12.1-q14.3 and gain of 13q33.3-qter. -
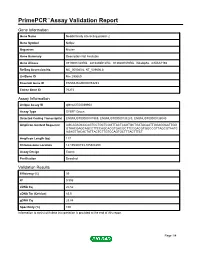
Primepcr™Assay Validation Report
PrimePCR™Assay Validation Report Gene Information Gene Name Nedd4 family interacting protein 2 Gene Symbol Ndfip2 Organism Mouse Gene Summary Description Not Available Gene Aliases 0710001O20Rik, 2810436B12Rik, 9130207N19Rik, N4wbp5a, mKIAA1165 RefSeq Accession No. NC_000080.6, NT_039606.8 UniGene ID Mm.290669 Ensembl Gene ID ENSMUSG00000053253 Entrez Gene ID 76273 Assay Information Unique Assay ID qMmuCED0039954 Assay Type SYBR® Green Detected Coding Transcript(s) ENSMUST00000181969, ENSMUST00000138283, ENSMUST00000136040 Amplicon Context Sequence AGCAGAGCCCAGTCCTGCTCGGTTACTCAGTGCTGATACAATTGGAGGAATTGG GTAACGAGCAGCCTTCCAGCACGTGACGCTTCCGACGTGGCCGTTAGCGTAATC AGAGTTACACTATTACTCTTGTCCAGTGCTTTACTTTCT Amplicon Length (bp) 117 Chromosome Location 14:105308153-105308299 Assay Design Exonic Purification Desalted Validation Results Efficiency (%) 99 R2 0.996 cDNA Cq 20.52 cDNA Tm (Celsius) 83.5 gDNA Cq 23.84 Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 1/4 PrimePCR™Assay Validation Report Ndfip2, Mouse Amplification Plot Amplification of cDNA generated from 25 ng of universal reference RNA Melt Peak Melt curve analysis of above amplification Standard Curve Standard curve generated using 20 million copies of template diluted 10-fold to 20 copies Page 2/4 PrimePCR™Assay Validation Report Products used to generate validation data Real-Time PCR Instrument CFX384 Real-Time PCR Detection System Reverse Transcription Reagent iScript™ Advanced cDNA Synthesis Kit for RT-qPCR Real-Time PCR Supermix SsoAdvanced™ SYBR® Green Supermix Experimental Sample qPCR Mouse Reference Total RNA Data Interpretation Unique Assay ID This is a unique identifier that can be used to identify the assay in the literature and online. Detected Coding Transcript(s) This is a list of the Ensembl transcript ID(s) that this assay will detect. -

Deletion of 11Q in Neuroblastomas Drives Sensitivity to PARP Inhibition Elena Sanmartín1,2, Lisandra Munoz~ 1,2, Marta Piqueras3, J
Published OnlineFirst August 22, 2017; DOI: 10.1158/1078-0432.CCR-17-0593 Personalized Medicine and Imaging Clinical Cancer Research Deletion of 11q in Neuroblastomas Drives Sensitivity to PARP Inhibition Elena Sanmartín1,2, Lisandra Munoz~ 1,2, Marta Piqueras3, J. Antoni Sirerol1,2, Pablo Berlanga2,4, Adela Canete~ 2,4, Victoria Castel2,4, and Jaime Font de Mora1,2 Abstract Purpose: Despite advances in multimodal therapy, neuroblas- define a subgroup of neuroblastomas with higher sensitivity to tomas with hemizygous deletion in chromosome 11q (20%– PARP inhibitors. Noteworthy, concomitant treatment with ola- 30%) undergo consecutive recurrences with poor outcome. We parib and DNA alkylating agent temozolomide potently inhibited hypothesized that patients with 11q-loss may share a druggable growth of cell lines harboring 11q-loss. This drug synergism was molecular target(s) that can be exploited for a precision medicine less potent when temozolomide was exchanged for cisplatin or strategy to improve treatment outcome. irinotecan. Intact 11q cells concomitantly treated with ATM Experimental Design: SNP arrays were combined with next- inhibitor displayed growth arrest and enhanced apoptosis, reveal- generation sequencing (NGS) to precisely define the deleted ing a role for ATM in the mechanism that mediates sensitivity to region in 17 primary 11q-loss neuroblastomas and identify allelic temozolomide–olaparib. Interestingly, functional TP53 is variants in genes relevant for neuroblastoma etiology. We required for efficacy of this treatment. In an in vivo model, assessed PARP inhibitor olaparib in combination with other coadministration of temozolomide–olaparib resulted in sus- chemotherapy medications using both in vitro and in vivo models. tained xenograft regression. -

Chloride Channels Regulate Differentiation and Barrier Functions
RESEARCH ARTICLE Chloride channels regulate differentiation and barrier functions of the mammalian airway Mu He1†*, Bing Wu2†, Wenlei Ye1, Daniel D Le2, Adriane W Sinclair3,4, Valeria Padovano5, Yuzhang Chen6, Ke-Xin Li1, Rene Sit2, Michelle Tan2, Michael J Caplan5, Norma Neff2, Yuh Nung Jan1,7,8, Spyros Darmanis2*, Lily Yeh Jan1,7,8* 1Department of Physiology, University of California, San Francisco, San Francisco, United States; 2Chan Zuckerberg Biohub, San Francisco, United States; 3Department of Urology, University of California, San Francisco, San Francisco, United States; 4Division of Pediatric Urology, University of California, San Francisco, Benioff Children’s Hospital, San Francisco, United States; 5Department of Cellular and Molecular Physiology, Yale University School of Medicine, New Heaven, United States; 6Department of Anesthesia and Perioperative Care, University of California, San Francisco, San Francisco, United States; 7Department of Biochemistry and Biophysics, University of California, San Francisco, San Francisco, United States; 8Howard Hughes Medical Institute, University of California, San Francisco, San Francisco, United States *For correspondence: Abstract The conducting airway forms a protective mucosal barrier and is the primary target of [email protected] (MH); [email protected] airway disorders. The molecular events required for the formation and function of the airway (SD); mucosal barrier, as well as the mechanisms by which barrier dysfunction leads to early onset airway [email protected] (LYJ) diseases, -

SOCS-Mediated Immunomodulation of Natural Killer Cells T ⁎ Narelle Keating, Sandra E
Cytokine 118 (2019) 64–70 Contents lists available at ScienceDirect Cytokine journal homepage: www.elsevier.com/locate/cytokine Review article SOCS-mediated immunomodulation of natural killer cells T ⁎ Narelle Keating, Sandra E. Nicholson Walter and Eliza Hall Institute of Medical Research, Melbourne 3052, Australia Department of Medical Biology, University of Melbourne, Melbourne 3010, Australia ARTICLE INFO ABSTRACT Keywords: Natural killer (NK) cells are innate immune cells with an intrinsic ability to detect and kill infected and can- Cancer cerous cells. The success of therapies targeting immune checkpoints on CD8 cells has intensified interest in Immune checkpoints harnessing the cytolytic effector functions of NK cells for new cancer treatments. NK cell development, survival NK cells and effector activity is dependent on exposure to the cytokine interleukin (IL)-15. The suppressor ofcytokine SOCS proteins (SOCS) proteins (CIS; SOCS1-7) are important negative regulators of cytokine signaling, and both CIS and SOCS2 CIS are reported to have roles in regulating NK cell responses. Their immunomodulatory effects on NK cells suggest SOCS2 that these SOCS proteins are promising targets that can potentially form the basis of novel cancer therapies. Here we discuss the role of NK cells in tumor immunity as well as review the role of the SOCS proteins in regulating IL- 15 signaling and NK cell function. 1. Introduction success of the immune checkpoint inhibitors has stimulated efforts to harness the anti-tumor effector functions of NK cells. Whilst CTLA-4 is Cancer is a major cause of disease burden worldwide, with an es- expressed on the surface of activated murine NK cells, it does not ap- timated 8.2 million cancer-related deaths in 2012 [1] and 1,735,000 pear to be expressed on human NK cells [6].