Strategies for Mapping and Cloning Quantitative Trait Genes in Rodents
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Murine MPDZ-Linked Hydrocephalus Is Caused by Hyperpermeability of the Choroid Plexus
Thomas Jefferson University Jefferson Digital Commons Cardeza Foundation for Hematologic Research Sidney Kimmel Medical College 1-1-2019 Murine MPDZ-linked hydrocephalus is caused by hyperpermeability of the choroid plexus. Junning Yang Thomas Jefferson University Claire Simonneau Thomas Jefferson University Robert Kilker Thomas Jefferson University Laura Oakley ThomasFollow this Jeff anderson additional University works at: https://jdc.jefferson.edu/cardeza_foundation Part of the Medical Pathology Commons Matthew D. Byrne ThomasLet us Jeff knowerson Univ howersity access to this document benefits ouy Recommended Citation See next page for additional authors Yang, Junning; Simonneau, Claire; Kilker, Robert; Oakley, Laura; Byrne, Matthew D.; Nichtova, Zuzana; Stefanescu, Ioana; Pardeep-Kumar, Fnu; Tripathi, Sushil; Londin, Eric; Saugier-Veber, Pascale; Willard, Belinda; Thakur, Mathew; Pickup, Stephen; Ishikawa, Hiroshi; Schroten, Horst; Smeyne, Richard; and Horowitz, Arie, "Murine MPDZ-linked hydrocephalus is caused by hyperpermeability of the choroid plexus." (2019). Cardeza Foundation for Hematologic Research. Paper 49. https://jdc.jefferson.edu/cardeza_foundation/49 This Article is brought to you for free and open access by the Jefferson Digital Commons. The Jefferson Digital Commons is a service of Thomas Jefferson University's Center for Teaching and Learning (CTL). The Commons is a showcase for Jefferson books and journals, peer-reviewed scholarly publications, unique historical collections from the University archives, and teaching tools. The Jefferson Digital Commons allows researchers and interested readers anywhere in the world to learn about and keep up to date with Jefferson scholarship. This article has been accepted for inclusion in Cardeza Foundation for Hematologic Research by an authorized administrator of the Jefferson Digital Commons. For more information, please contact: [email protected]. -

Lineage-Specific Mapping of Quantitative Trait Loci
Heredity (2013) 111, 106–113 & 2013 Macmillan Publishers Limited All rights reserved 0018-067X/13 www.nature.com/hdy ORIGINAL ARTICLE Lineage-specific mapping of quantitative trait loci C Chen1 and K Ritland We present an approach for quantitative trait locus (QTL) mapping, termed as ‘lineage-specific QTL mapping’, for inferring allelic changes of QTL evolution along with branches in a phylogeny. We describe and analyze the simplest case: by adding a third taxon into the normal procedure of QTL mapping between pairs of taxa, such inferences can be made along lineages to a presumed common ancestor. Although comparisons of QTL maps among species can identify homology of QTLs by apparent co- location, lineage-specific mapping of QTL can classify homology into (1) orthology (shared origin of QTL) versus (2) paralogy (independent origin of QTL within resolution of map distance). In this light, we present a graphical method that identifies six modes of QTL evolution in a three taxon comparison. We then apply our model to map lineage-specific QTLs for inbreeding among three taxa of yellow monkey-flower: Mimulus guttatus and two inbreeders M. platycalyx and M. micranthus, but critically assuming outcrossing was the ancestral state. The two most common modes of homology across traits were orthologous (shared ancestry of mutation for QTL alleles). The outbreeder M. guttatus had the fewest lineage-specific QTL, in accordance with the presumed ancestry of outbreeding. Extensions of lineage-specific QTL mapping to other types of data and crosses, and to inference of ancestral QTL state, are discussed. Heredity (2013) 111, 106–113; doi:10.1038/hdy.2013.24; published online 24 April 2013 Keywords: Quantitative trait locus mapping; evolution of mating systems; Mimulus INTRODUCTION common ancestor of these two taxa, back to the common ancestor of Knowledge of the genetic architecture of complex traits can offer all three taxa and forwards again to the third taxon. -
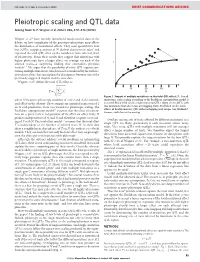
Pleiotropic Scaling and QTL Data Arising From: G
NATURE | Vol 456 | 4 December 2008 BRIEF COMMUNICATIONS ARISING Pleiotropic scaling and QTL data Arising from: G. P. Wagner et al. Nature 452, 470–473 (2008) Wagner et al.1 have recently introduced much-needed data to the debate on how complexity of the genotype–phenotype map affects the distribution of mutational effects. They used quantitative trait loci (QTLs) mapping analysis of 70 skeletal characters in mice2 and regressed the total QTL effect on the number of traits affected (level of pleiotropy). From their results they suggest that mutations with higher pleiotropy have a larger effect, on average, on each of the T affected traits—a surprising finding that contradicts previous models3–7. We argue that the possibility of some QTL regions con- taining multiple mutations, which was not considered by the authors, introduces a bias that can explain the discrepancy between one of the previously suggested models and the new data. Wagner et al.1 define the total QTL effect as vffiffiffiffiffiffiffiffiffiffiffiffiffi u uXN 10 20 30 ~t 2 T Ai N i~1 Figure 1 | Impact of multiple mutations on the total QTL effect, T. For all where N measures pleiotropy (number of traits) and Ai the normal- mutations, strict scaling according to the Euclidean superposition model is ized effect on the ith trait. They compare an empirical regression of T assumed. Black filled circles, single-mutation QTLs. Open circles, QTLs with on N with predictions from two models for pleiotropic scaling. The two mutations that affect non-overlapping traits. Red filled circles, total Euclidean superposition model3,4 assumes that the effect of a muta- effects of double-mutant QTL with overlapping trait ranges (see Methods). -

Roles and Mechanisms of Kinesin-6 KIF20A in Spindle Organization During Cell Division T ⁎ Wen-Da Wu, Kai-Wei Yu, Ning Zhong, Yu Xiao, Zhen-Yu She
European Journal of Cell Biology 98 (2019) 74–80 Contents lists available at ScienceDirect European Journal of Cell Biology journal homepage: www.elsevier.com/locate/ejcb Review Roles and mechanisms of Kinesin-6 KIF20A in spindle organization during cell division T ⁎ Wen-Da Wu, Kai-Wei Yu, Ning Zhong, Yu Xiao, Zhen-Yu She Department of Cell Biology and Genetics/Center for Cell and Developmental Biology, The School of Basic Medical Sciences, Fujian Medical University, Fuzhou, Fujian 350108, China ARTICLE INFO ABSTRACT Keywords: Mitotic kinesin is crucial for spindle assembly and chromosome segregation in cell division. KIF20A/MKlp2, a Kinesin-6 member of kinesin-6 subfamily, plays important roles in the central spindle organization at anaphase and cy- KIF20A tokinesis. In this review, we briefly introduce the discovery and classification of kinesin-6 motors in model Microtubule organisms, and summarize the biochemical features and mechanics of KIF20A proteins. We emphasize the Anaphase complicated interactions of KIF20A with partner proteins, including MKlp1, Plk1 and Rab6. Particularly, we Spindle assembly highlight the regulation of Cdk1 and chromosomal passenger complex on kinesin-6 KIF20A at late stage of Mitosis mitosis. We summarized the multiple functions of KIF20A in central spindle assembly and the formation of cleavage furrow in both mitosis and meiosis. In addition, we conclude the expression patterns of KIF20A in tumorigenesis and its applications in tumor therapy. 1. Introduction kinesin superfamily proteins (Miki et al., 2005). Kinesin-6 subfamily is comprised of KIF20A (Lawrence et al., 2004), KIF20B (MPP1) Kinesin superfamily proteins (KIFs) are molecular motors that (Kamimoto et al., 2001; Matsumoto-Taniura et al., 1996; Westendorf mediate the transport of various cargos, including the newly synthe- et al., 1994) and MKlp1 (Lawrence et al., 2004; Nislow et al., 1990; sized protein complexes, vesicles and mRNAs along the microtubule Sellitto and Kuriyama, 1988). -

The Utility of Genetic Risk Scores in Predicting the Onset of Stroke March 2021 6
DOT/FAA/AM-21/24 Office of Aerospace Medicine Washington, DC 20591 The Utility of Genetic Risk Scores in Predicting the Onset of Stroke Diana Judith Monroy Rios, M.D1 and Scott J. Nicholson, Ph.D.2 1. KR 30 # 45-03 University Campus, Building 471, 5th Floor, Office 510 Bogotá D.C. Colombia 2. FAA Civil Aerospace Medical Institute, 6500 S. MacArthur Blvd Rm. 354, Oklahoma City, OK 73125 March 2021 NOTICE This document is disseminated under the sponsorship of the U.S. Department of Transportation in the interest of information exchange. The United States Government assumes no liability for the contents thereof. _________________ This publication and all Office of Aerospace Medicine technical reports are available in full-text from the Civil Aerospace Medical Institute’s publications Web site: (www.faa.gov/go/oamtechreports) Technical Report Documentation Page 1. Report No. 2. Government Accession No. 3. Recipient's Catalog No. DOT/FAA/AM-21/24 4. Title and Subtitle 5. Report Date March 2021 The Utility of Genetic Risk Scores in Predicting the Onset of Stroke 6. Performing Organization Code 7. Author(s) 8. Performing Organization Report No. Diana Judith Monroy Rios M.D1, and Scott J. Nicholson, Ph.D.2 9. Performing Organization Name and Address 10. Work Unit No. (TRAIS) 1 KR 30 # 45-03 University Campus, Building 471, 5th Floor, Office 510, Bogotá D.C. Colombia 11. Contract or Grant No. 2 FAA Civil Aerospace Medical Institute, 6500 S. MacArthur Blvd Rm. 354, Oklahoma City, OK 73125 12. Sponsoring Agency name and Address 13. Type of Report and Period Covered Office of Aerospace Medicine Federal Aviation Administration 800 Independence Ave., S.W. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

1 ROBERT PLOMIN on Behavioral Genetics Genetic Links Common
Genetic links • Background – Why has educational psychology ben so slow ROBERT PLOMIN to accept genetics? on Behavioral Genetics • Beyond nature vs. nurture – 3 findings from genetic research with far- reaching implications for education: October, 2004 The nature of nurture The abnormal is normal Generalist genes • Molecular Genetics Quantitative genetics: twin and adoption studies Common medical disorders Identical twins Fraternal twins (monozygotic, MZ) (dizygotic, DZ) 1 Twin concordances in TEDS at 7 years Twins Early Development Study (TEDS) for low reading and maths Perceptions of nature/nurture: % parents (N=1340) and teachers Beyond nature vs. nurture: (N=667) indicating that genes are at least as important as environment (Walker & Plomin, Ed. Psych., in press) The nature of nurture • Genetic influence on experience • Genotype-environment correlation – Children’s genetic propensities are correlated with their experiences passive evocative active 2 Beyond nature vs. nurture: Genotype-environment correlation The nature of nurture • Passive • Genotype-environment correlation – e.g., family background (SES) and school – Children’s genetic propensities are correlated with achievement their experiences passive • Evocative evocative – Children evoke experiences for genetic reasons Active • Active • Molecular genetics – Children actively create environments that foster – Find genes associated with learning experiences their genetic propensities Beyond nature vs. nurture: The one-gene one-disability (OGOD) model The abnormal is normal • Common learning disabilities are the quantitative extreme of the same genetic and environmental factors responsible for normal variation in learning abilities. • There are no common learning disabilities, just the extremes of normal variation. 3 OGOD (rare) and QTL (common) disorders The quantitative trait locus (QTL) model Beyond nature vs. -

Supplementary Table 1: Adhesion Genes Data Set
Supplementary Table 1: Adhesion genes data set PROBE Entrez Gene ID Celera Gene ID Gene_Symbol Gene_Name 160832 1 hCG201364.3 A1BG alpha-1-B glycoprotein 223658 1 hCG201364.3 A1BG alpha-1-B glycoprotein 212988 102 hCG40040.3 ADAM10 ADAM metallopeptidase domain 10 133411 4185 hCG28232.2 ADAM11 ADAM metallopeptidase domain 11 110695 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 195222 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 165344 8751 hCG20021.3 ADAM15 ADAM metallopeptidase domain 15 (metargidin) 189065 6868 null ADAM17 ADAM metallopeptidase domain 17 (tumor necrosis factor, alpha, converting enzyme) 108119 8728 hCG15398.4 ADAM19 ADAM metallopeptidase domain 19 (meltrin beta) 117763 8748 hCG20675.3 ADAM20 ADAM metallopeptidase domain 20 126448 8747 hCG1785634.2 ADAM21 ADAM metallopeptidase domain 21 208981 8747 hCG1785634.2|hCG2042897 ADAM21 ADAM metallopeptidase domain 21 180903 53616 hCG17212.4 ADAM22 ADAM metallopeptidase domain 22 177272 8745 hCG1811623.1 ADAM23 ADAM metallopeptidase domain 23 102384 10863 hCG1818505.1 ADAM28 ADAM metallopeptidase domain 28 119968 11086 hCG1786734.2 ADAM29 ADAM metallopeptidase domain 29 205542 11085 hCG1997196.1 ADAM30 ADAM metallopeptidase domain 30 148417 80332 hCG39255.4 ADAM33 ADAM metallopeptidase domain 33 140492 8756 hCG1789002.2 ADAM7 ADAM metallopeptidase domain 7 122603 101 hCG1816947.1 ADAM8 ADAM metallopeptidase domain 8 183965 8754 hCG1996391 ADAM9 ADAM metallopeptidase domain 9 (meltrin gamma) 129974 27299 hCG15447.3 ADAMDEC1 ADAM-like, -

Epigenome-Wide Study Identified Methylation Sites Associated With
nutrients Article Epigenome-Wide Study Identified Methylation Sites Associated with the Risk of Obesity Majid Nikpay 1,*, Sepehr Ravati 2, Robert Dent 3 and Ruth McPherson 1,4,* 1 Ruddy Canadian Cardiovascular Genetics Centre, University of Ottawa Heart Institute, 40 Ruskin St–H4208, Ottawa, ON K1Y 4W7, Canada 2 Plastenor Technologies Company, Montreal, QC H2P 2G4, Canada; [email protected] 3 Department of Medicine, Division of Endocrinology, University of Ottawa, the Ottawa Hospital, Ottawa, ON K1Y 4E9, Canada; [email protected] 4 Atherogenomics Laboratory, University of Ottawa Heart Institute, Ottawa, ON K1Y 4W7, Canada * Correspondence: [email protected] (M.N.); [email protected] (R.M.) Abstract: Here, we performed a genome-wide search for methylation sites that contribute to the risk of obesity. We integrated methylation quantitative trait locus (mQTL) data with BMI GWAS information through a SNP-based multiomics approach to identify genomic regions where mQTLs for a methylation site co-localize with obesity risk SNPs. We then tested whether the identified site contributed to BMI through Mendelian randomization. We identified multiple methylation sites causally contributing to the risk of obesity. We validated these findings through a replication stage. By integrating expression quantitative trait locus (eQTL) data, we noted that lower methylation at cg21178254 site upstream of CCNL1 contributes to obesity by increasing the expression of this gene. Higher methylation at cg02814054 increases the risk of obesity by lowering the expression of MAST3, whereas lower methylation at cg06028605 contributes to obesity by decreasing the expression of SLC5A11. Finally, we noted that rare variants within 2p23.3 impact obesity by making the cg01884057 Citation: Nikpay, M.; Ravati, S.; Dent, R.; McPherson, R. -

High Throughput Computational Mouse Genetic Analysis
bioRxiv preprint doi: https://doi.org/10.1101/2020.09.01.278465; this version posted January 22, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. High Throughput Computational Mouse Genetic Analysis Ahmed Arslan1+, Yuan Guan1+, Zhuoqing Fang1, Xinyu Chen1, Robin Donaldson2&, Wan Zhu, Madeline Ford1, Manhong Wu, Ming Zheng1, David L. Dill2* and Gary Peltz1* 1Department of Anesthesia, Stanford University School of Medicine, Stanford, CA; and 2Department of Computer Science, Stanford University, Stanford, CA +These authors contributed equally to this paper & Current Address: Ecree Durham, NC 27701 *Address correspondence to: [email protected] bioRxiv preprint doi: https://doi.org/10.1101/2020.09.01.278465; this version posted January 22, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract Background: Genetic factors affecting multiple biomedical traits in mice have been identified when GWAS data that measured responses in panels of inbred mouse strains was analyzed using haplotype-based computational genetic mapping (HBCGM). Although this method was previously used to analyze one dataset at a time; but now, a vast amount of mouse phenotypic data is now publicly available, which could lead to many more genetic discoveries. Results: HBCGM and a whole genome SNP map covering 53 inbred strains was used to analyze 8462 publicly available datasets of biomedical responses (1.52M individual datapoints) measured in panels of inbred mouse strains. As proof of concept, causative genetic factors affecting susceptibility for eye, metabolic and infectious diseases were identified when structured automated methods were used to analyze the output. -

QUANTITATIVE TRAIT LOCUS ANALYSIS: in Several Variants (I.E., Alleles)
TECHNOLOGIES FROM THE FIELD QUANTITATIVE TRAIT LOCUS ANALYSIS: in several variants (i.e., alleles). QTL analysis allows one MULTIPLE CROSS AND HETEROGENEOUS to map, with some precision, the genomic position of STOCK MAPPING these alleles. For many researchers in the alcohol field, the break Robert Hitzemann, Ph.D.; John K. Belknap, Ph.D.; through with respect to the genetic determination of alcohol- and Shannon K. McWeeney, Ph.D. related behaviors occurred when Plomin and colleagues (1991) made the seminal observation that a specific panel of RI mice (i.e., the BXD panel) could be used to identify KEY WORDS: Genetic theory of alcohol and other drug use; genetic the physical location of (i.e., to map) QTLs for behavioral factors; environmental factors; behavioral phenotype; behavioral phenotypes. Because the phenotypes of the different strains trait; quantitative trait gene (QTG); inbred animal strains; in this panel had been determined for many alcohol- recombinant inbred (RI) mouse strains; quantitative traits; related traits, researchers could readily apply the strategy quantitative trait locus (QTL) mapping; multiple cross mapping of RI–QTL mapping (Gora-Maslak et al. 1991). Although (MCM); heterogeneous stock (HS) mapping; microsatellite investigators recognized early on that this panel was not mapping; animal models extensive enough to answer all questions, the emerging data illustrated the rich genetic complexity of alcohol- related phenotypes (Belknap 1992; Plomin and McClearn ntil well into the 1990s, both preclinical and clinical 1993). research focused on finding “the” gene for human The next advance came with the development of diseases, including alcoholism. This focus was rein U microsatellite maps (Dietrich et al. -

Organ of Corti Size Is Governed by Yap/Tead-Mediated Progenitor Self-Renewal
Organ of Corti size is governed by Yap/Tead-mediated progenitor self-renewal Ksenia Gnedevaa,b,1, Xizi Wanga,b, Melissa M. McGovernc, Matthew Bartond,2, Litao Taoa,b, Talon Treceka,b, Tanner O. Monroee,f, Juan Llamasa,b, Welly Makmuraa,b, James F. Martinf,g,h, Andrew K. Grovesc,g,i, Mark Warchold, and Neil Segila,b,1 aDepartment of Stem Cell Biology and Regenerative Medicine, Keck Medicine of University of Southern California, Los Angeles, CA 90033; bCaruso Department of Otolaryngology–Head and Neck Surgery, Keck Medicine of University of Southern California, Los Angeles, CA 90033; cDepartment of Neuroscience, Baylor College of Medicine, Houston, TX 77030; dDepartment of Otolaryngology, Washington University in St. Louis, St. Louis, MO 63130; eAdvanced Center for Translational and Genetic Medicine, Lurie Children’s Hospital of Chicago, Chicago, IL 60611; fDepartment of Molecular Physiology and Biophysics, Baylor College of Medicine, Houston, TX 77030; gProgram in Developmental Biology, Baylor College of Medicine, Houston, TX 77030; hCardiomyocyte Renewal Laboratory, Texas Heart Institute, Houston, TX 77030 and iDepartment of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX 77030; Edited by Marianne E. Bronner, California Institute of Technology, Pasadena, CA, and approved April 21, 2020 (received for review January 6, 2020) Precise control of organ growth and patterning is executed However, what initiates this increase in Cdkn1b expression re- through a balanced regulation of progenitor self-renewal and dif- mains unclear. In addition, conditional ablation of Cdkn1b in the ferentiation. In the auditory sensory epithelium—the organ of inner ear is not sufficient to completely relieve the block on Corti—progenitor cells exit the cell cycle in a coordinated wave supporting cell proliferation (9, 10), suggesting the existence of between E12.5 and E14.5 before the initiation of sensory receptor additional repressive mechanisms.