Electronic Supplementary Material Evolutionary Rates Are Correlated
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

New Aspects About Supella Longipalpa (Blattaria: Blattellidae)
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector Asian Pac J Trop Biomed 2016; 6(12): 1065–1075 1065 HOSTED BY Contents lists available at ScienceDirect Asian Pacific Journal of Tropical Biomedicine journal homepage: www.elsevier.com/locate/apjtb Review article http://dx.doi.org/10.1016/j.apjtb.2016.08.017 New aspects about Supella longipalpa (Blattaria: Blattellidae) Hassan Nasirian* Department of Medical Entomology and Vector Control, School of Public Health, Tehran University of Medical Sciences, Tehran, Iran ARTICLE INFO ABSTRACT Article history: The brown-banded cockroach, Supella longipalpa (Blattaria: Blattellidae) (S. longipalpa), Received 16 Jun 2015 recently has infested the buildings and hospitals in wide areas of Iran, and this review was Received in revised form 3 Jul 2015, prepared to identify current knowledge and knowledge gaps about the brown-banded 2nd revised form 7 Jun, 3rd revised cockroach. Scientific reports and peer-reviewed papers concerning S. longipalpa and form 18 Jul 2016 relevant topics were collected and synthesized with the objective of learning more about Accepted 10 Aug 2016 health-related impacts and possible management of S. longipalpa in Iran. Like the Available online 15 Oct 2016 German cockroach, the brown-banded cockroach is a known vector for food-borne dis- eases and drug resistant bacteria, contaminated by infectious disease agents, involved in human intestinal parasites and is the intermediate host of Trichospirura leptostoma and Keywords: Moniliformis moniliformis. Because its habitat is widespread, distributed throughout Brown-banded cockroach different areas of homes and buildings, it is difficult to control. -

Toxicological Characteristics of Edible Insects in China: a Historical Review
Accepted Manuscript Toxicological characteristics of edible insects in China: A historical review Yu Gao, Di Wang, Meng-Lei Xu, Shu-Sen Shi, Jin-Feng Xiong PII: S0278-6915(18)30218-7 DOI: 10.1016/j.fct.2018.04.016 Reference: FCT 9705 To appear in: Food and Chemical Toxicology Received Date: 26 January 2018 Revised Date: 1 April 2018 Accepted Date: 7 April 2018 Please cite this article as: Gao, Y., Wang, D., Xu, M.-L., Shi, S.-S., Xiong, J.-F., Toxicological characteristics of edible insects in China: A historical review, Food and Chemical Toxicology (2018), doi: 10.1016/j.fct.2018.04.016. This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain. ACCEPTED MANUSCRIPT 1 2 Toxicological Characteristics of Edible Insects in China: A historical review 3 Yu Gao a, Di Wang a, Meng-Lei Xu b*, Shu-Sen Shi a* , Jin-Feng Xiong c 4 5 a College of Agriculture, Jilin Agricultural University, Changchun, 130118, P. R. 6 China 7 b State Key Laboratory of Supramolecular Structure and Materials, Jilin University, 8 Changchun, 130000, P. R. China 9 c Changchun Institute of Biological Products Co. Ltd., Changchun, 130012, P. R. -

Cockroach Marion Copeland
Cockroach Marion Copeland Animal series Cockroach Animal Series editor: Jonathan Burt Already published Crow Boria Sax Tortoise Peter Young Ant Charlotte Sleigh Forthcoming Wolf Falcon Garry Marvin Helen Macdonald Bear Parrot Robert E. Bieder Paul Carter Horse Whale Sarah Wintle Joseph Roman Spider Rat Leslie Dick Jonathan Burt Dog Hare Susan McHugh Simon Carnell Snake Bee Drake Stutesman Claire Preston Oyster Rebecca Stott Cockroach Marion Copeland reaktion books Published by reaktion books ltd 79 Farringdon Road London ec1m 3ju, uk www.reaktionbooks.co.uk First published 2003 Copyright © Marion Copeland All rights reserved No part of this publication may be reproduced, stored in a retrieval system or transmitted, in any form or by any means, electronic, mechanical, photocopying, recording or otherwise without the prior permission of the publishers. Printed and bound in Hong Kong British Library Cataloguing in Publication Data Copeland, Marion Cockroach. – (Animal) 1. Cockroaches 2. Animals and civilization I. Title 595.7’28 isbn 1 86189 192 x Contents Introduction 7 1 A Living Fossil 15 2 What’s in a Name? 44 3 Fellow Traveller 60 4 In the Mind of Man: Myth, Folklore and the Arts 79 5 Tales from the Underside 107 6 Robo-roach 130 7 The Golden Cockroach 148 Timeline 170 Appendix: ‘La Cucaracha’ 172 References 174 Bibliography 186 Associations 189 Websites 190 Acknowledgements 191 Photo Acknowledgements 193 Index 196 Two types of cockroach, from the first major work of American natural history, published in 1747. Introduction The cockroach could not have scuttled along, almost unchanged, for over three hundred million years – some two hundred and ninety-nine million before man evolved – unless it was doing something right. -

Bulletin Number / Numéro 2 Entomological Society of Canada Société D’Entomologie Du Canada June / Juin 2008
Volume 40 Bulletin Number / numéro 2 Entomological Society of Canada Société d’entomologie du Canada June / juin 2008 Published quarterly by the Entomological Society of Canada Publication trimestrielle par la Société d’entomologie du Canada ............................................................... .................................................................................................................................................................................................................................................................................................................................. .......................................................................... ........................................................................................................................................................................ ....................... ................................................................................. ................................................. List of contents / Table des matières Volume 40 (2), June / june 2008 Up front / Avant-propos ................................................................................................................49 Moth balls / Boules à mites .............................................................................................................51 Meeting announcements / Réunions futures ..................................................................................52 Dear Buggy / Cher Bibitte ..............................................................................................................53 -
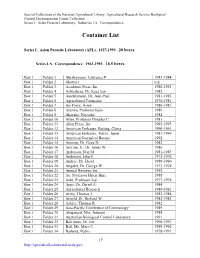
Container List
Special Collections of the National Agricultural Library: Agricultural Research Service Biological Control Documentation Center Collection Series I. Asian Parasite Laboratory. Subseries I.A. Correspondence. Container List Series I. Asian Parasite Laboratory (APL). 1927-1993. 20 boxes. Series I.A. Correspondence. 1963-1993. 16.5 boxes. Box 1 Folder 1 Abrahamson, Lawrence P. 1983-1984 Box 1 Folder 2 Abstract n.d. Box 1 Folder 3 Academic Press, Inc. 1986-1993 Box 1 Folder 4 Achterberg, Dr. Kees van 1982 Box 1 Folder 5 Aeschlimann, Dr. Jean-Paul 1981-1985 Box 1 Folder 6 Agricultural Counselor 1976-1981 Box 1 Folder 7 Air Force, Army 1980-1981 Box 1 Folder 8 Aizawa, Professor Keio 1986 Box 1 Folder 9 Akasaka, Naoyuki 1982 Box 1 Folder 10 Allen, Professor Douglas C. 1981 Box 1 Folder 11 Allen Press, Inc. 1992-1993 Box 1 Folder 12 American Embassy, Beijing, China 1990-1991 Box 1 Folder 13 American Embassy, Tokyo, Japan 1981-1990 Box 1 Folder 14 American Journal of Botany 1992 Box 1 Folder 15 Amman, Dr. Gene D. 1982 Box 1 Folder 16 Amrine, Jr., Dr. James W. 1986 Box 1 Folder 17 Anderson, Don M. 1981-1983 Box 1 Folder 18 Anderson, John F. 1975-1976 Box 1 Folder 19 Andres, Dr. Lloyd 1989-1990 Box 1 Folder 20 Angalet, Dr. George W. 1973-1978 Box 1 Folder 21 Annual Reviews Inc. 1992 Box 1 Folder 22 Ao, Professor Hsien-Bine 1988 Box 1 Folder 23 Aoki, Professor Joji 1977-1978 Box 1 Folder 24 Apps, Dr. Darrel A. 1984 Box 1 Folder 25 Agricultural Research 1980-1981 Box 1 Folder 26 Army, Thomas J. -

RESEARCH ARTICLE a New Species of Cockroach, Periplaneta
Tropical Biomedicine 38(2): 48-52 (2021) https://doi.org/10.47665/tb.38.2.036 RESEARCH ARTICLE A new species of cockroach, Periplaneta gajajimana sp. nov., collected in Gajajima, Kagoshima Prefecture, Japan Komatsu, N.1, Iio, H.2, Ooi, H.K.3* 1Civil International Corporation, 10–14 Kitaueno 1, Taito–ku, Tokyo, 110–0014, Japan 2Foundation for the Protection of Deer in Nara, 160-1 Kasugano-cho, Nara-City, Nara, 630-8212, Japan 3Laboratory of Parasitology, School of Veterinary Medicine, Azabu University, 1-17-710 Fuchinobe, Sagamihara, Kanagawa 252-5201 Japan *Corresponding author: [email protected] ARTICLE HISTORY ABSTRACT Received: 25 January 2021 We described a new species of cockroach, Periplaneta gajajimana sp. nov., which was collected Revised: 2 February 2021 in Gajajima, Kagoshima-gun Toshimamura, Kagoshima Prefecture, Japan, on November 2012. Accepted: 2 February 2021 The new species is characterized by its reddish brown to blackish brown body, smooth Published: 30 April 2021 surface pronotum, well developed compound eyes, dark brown head apex, dark reddish brown front face and small white ocelli connected to the antennal sockets. In male, the tegmen tip reach the abdomen end or are slightly shorter, while in the female, it does not reach the abdominal end and exposes the abdomen beyond the 7th abdominal plate. We confirmed the validity of this new species by breeding the specimens in our laboratory to demonstrate that the features of the progeny were maintained for several generations. For comparison and easy identification of this new species, the key to species identification of the genus Periplaneta that had been reported in Japan to date are also presented. -

INSECTA: Blattodea, Ectobiidae)*
BOLETÍN CIENTÍFICO ISSN 0123 - 3068 bol.cient.mus.hist.nat. 16 (2): 185 - 197 CENTRO DE MUSEOS MUSEO DE HISTORIA NATURAL RELACIÓN E ILUSTRACIÓN DE ALGUNAS ESPECIES DE NYCTIBORINAE DE COLOMBIA Y COSTA RICA (INSECTA: Blattodea, Ectobiidae)* Julián A. Salazar- E1- J. Cristóbal Ríos Maláver2 Resumen Se ilustran a color ejemplares de algunos Nyctiborinae neotropicales, con la figuración de los tipos de 3 especies de esta subfamilia, descritos de Colombia y Costa Rica: Paratropes otunensis Salazar, 2004; Muzoa simplex Hebard, 1921 y Muzoa madida Rhen, 1930. Además se incluyen caracteres relevantes de los principales géneros registrados en Colombia, una clave dicotómica a nivel genérico e ilustración de especies procedentes de algunas colecciones nacionales. Tales especies fueron originalmente descritas y publicadas en diversas revistas nacionales o internacionales. Se busca ante todo ofrecer información sobre su taxonomía e ilustraciones de buena calidad de espécimenes preservados que faciliten su identidad y fácil reconocimiento. La información incluye el nombre científico completo, los nombres de los autores, la publicación específica y el año de descripción. Además en lo posible la citación de material adicional perteneciente de cada especie tratada. Palabras clave: Holotipos, material, Blattodea, Nyctiborinae, Colombia, Costa Rica, neotrópico RELATION AND ILLUSTRATION OF SOME NYCTIBORINAE SPECIES FROM COLOMBIA AND COSTA RICA (INSECTA: Blattodea, Ectobiidae) Abstract Specimens of some neotropical Nyctiborinae are illustrated in full color with the inclusion of three species of this subfamily known from Colombia and Costa Rica: Paratropes otunensis Salazar, 2004; Muzoa simplex Hebard, 1921 and Muzoa madida Rhen, 1930. Also relevant characters from the main genera registered in Colombia are included, a dycotomic key at the genera and illustration level of species coming from some national collections. -

Pennsylvania Schools and Childcares Contents
For PennsylvaniaIPM Schools and Childcares A HOW-TO MANUAL Introduction to the 2019 Edition The first edition of this manual was published in importance of “good bugs” and the need for 2002. Shortly thereafter legislation was enacted conservation and provides tips for attracting requiring each Pennsylvania school district, beneficial organisms to school and childcare intermediate unit, and area vocational-technical gardens and grounds. school to develop an integrated pest management • The addition of “How to Develop an IPM (IPM) plan (Act 35 of 2002), notify parents and Policy and Plan for Your School District or guardians 72 hours prior to any pesticide appli- Childcare Facility/Provider” on page 15 shows cations, and post warning signs 72 hours prior to the parts of an acceptable plan with an and 48 hours after any pesticide applications in outline to adapt it for your school/childcare. school buildings or on school grounds (Act 36 of 2002). • The Pennsylvania School Boards Association In 2012, the Pennsylvania Department of policy has been replaced with an updated Agriculture (PDA) Health and Safety Division version reflecting the effects of the school determined that these regulations apply to child- IPM legislation. cares as well as K–12 schools since childcare facil- ities are explicitly covered by the Pennsylvania • A sample notification letter for parents and Pesticide Control Act of 1973. Note that this guardians has been added, as well as a includes the provision that only a licensed pest sample pest control information sheet used control operator can apply a pesticide in a facility to inform staff and parents and guardians or a home-based childcare center. -

A Dichotomous Key for the Identification of the Cockroach Fauna (Insecta: Blattaria) of Florida
Species Identification - Cockroaches of Florida 1 A Dichotomous Key for the Identification of the Cockroach fauna (Insecta: Blattaria) of Florida Insect Classification Exercise Department of Entomology and Nematology University of Florida, Gainesville 32611 Abstract: Students used available literature and specimens to produce a dichotomous key to species of cockroaches recorded from Florida. This exercise introduced students to techniques used in studying a group of insects, in this case Blattaria, to produce a regional species key. Producing a guide to a group of insects as a class exercise has proven useful both as a teaching tool and as a method to generate information for the public. Key Words: Blattaria, Florida, Blatta, Eurycotis, Periplaneta, Arenivaga, Compsodes, Holocompsa, Myrmecoblatta, Blatella, Cariblatta, Chorisoneura, Euthlastoblatta, Ischnoptera,Latiblatta, Neoblatella, Parcoblatta, Plectoptera, Supella, Symploce,Blaberus, Epilampra, Hemiblabera, Nauphoeta, Panchlora, Phoetalia, Pycnoscelis, Rhyparobia, distributions, systematics, education, teaching, techniques. Identification of cockroaches is limited here to adults. A major source of confusion is the recogni- tion of adults from nymphs (Figs. 1, 2). There are subjective differences, as well as morphological differences. Immature cockroaches are known as nymphs. Nymphs closely resemble adults except nymphs are generally smaller and lack wings and genital openings or copulatory appendages at the tip of their abdomen. Many species, however, have wingless adult females. Nymphs of these may be recognized by their shorter, relatively broad cerci and lack of external genitalia. Male cockroaches possess styli in addition to paired cerci. Styli arise from the subgenital plate and are generally con- spicuous, but may also be reduced in some species. Styli are absent in adult females and nymphs. -

Towards Classical Biological Control of Leek Moth
____________________________________________________________________________ Ateyyat This project seeks to provide greater coherence for the biocontrol knowledge system for regulators and researchers; create an open access information source for biocontrol re- search of agricultural pests in California, which will stimulate greater international knowl- edge sharing about agricultural pests in Mediterranean climates; and facilitate the exchange of information through a cyberinfrastructure among government regulators, and biocontrol entomologists and practitioners. It seeks broader impacts through: the uploading of previ- ously unavailable data being made openly accessible; the stimulation of greater interaction between the biological control regulation, research, and practitioner community in selected Mediterranean regions; the provision of more coherent and useful information to enhance regulatory decisions by public agency scientists; a partnership with the IOBC to facilitate international data sharing; and progress toward the ultimate goal of increasing the viability of biocontrol as a reduced risk pest control strategy. No Designated Session Theme BIOLOGY OF CIRROSPILUS INGENUUS GAHAN (HYMENOPTERA: EULOPHIDAE), AN ECTOPARASITOID OF THE CITRUS LEAFMINER, PHYLLOCNISTIS CITRELLA STAINTON (LEPIDOPTERA: GRACILLARIIDAE) ON LEMON 99 Mazen A. ATEYYAT Al-Shoubak University College, Al-Balqa’ Applied University, P.O. Box (5), Postal code 71911, Al-Shawbak, Jordan [email protected] The citrus leafminer (CLM), Phyllocnistis citrella Stainton (Lepidoptera: Gracillariidae) in- vaded the Jordan Valley in 1994 and was able to spread throughout Jordan within a few months of its arrival. It was the most common parasitoid from 1997 to 1999 in the Jordan Valley. An increase in the activity of C. ingenuus was observed in autumn and the highest number of emerged C. ingenuus adults was in November 1999. -

Cockroaches Introduction Members of the Order Blattodea, There Are About 4,500 Species of Cockroach, Which Live in a Wide Range of Environments Around the World
The following sample is taken from Breeding Insects as Feeder Food published as both an ebook and in hardcopy on Amazon. The images have been removed from this sample but the formatting is retained. Cockroaches Introduction Members of the order Blattodea, there are about 4,500 species of cockroach, which live in a wide range of environments around the world. Although considered pests associated with disease, there are only about 30 species that are commonly found in proximity with humans and, of these, only four are considered serious pests. Cockroaches are mainly nocturnal and will normally try to avoid light. Although most species prefer warm climates, cockroaches are among the hardiest insects and can survive extremes at both end of the temperature scale. Some species are capable of remaining active for a month without food and are able to survive on very limited sustenance. They are omnivorous, feeding on organic materials, including decaying plant matter, but they will also eat dead insects and animals. Compared to crickets, cockroaches have some advantages as feeder insects. They are unlikely to attack a small chameleon in the same way that a cricket might. They are longer lived and are hardier. Most are easier to breed than crickets and they make no noise! Unfortunately, despite these advantages, they are not as readily accepted by some animals as crickets are. Cockroaches suffer from a perception they harbour disease and that they smell. The species shown below will not do so if kept in clean conditions and fed correctly. For mantis, cockroaches can be a better choice than crickets. -

Thesis (PDF, 13.51MB)
Insects and their endosymbionts: phylogenetics and evolutionary rates Daej A Kh A M Arab The University of Sydney Faculty of Science 2021 A thesis submitted in fulfilment of the requirements for the degree of Doctor of Philosophy Authorship contribution statement During my doctoral candidature I published as first-author or co-author three stand-alone papers in peer-reviewed, internationally recognised journals. These publications form the three research chapters of this thesis in accordance with The University of Sydney’s policy for doctoral theses. These chapters are linked by the use of the latest phylogenetic and molecular evolutionary techniques for analysing obligate mutualistic endosymbionts and their host mitochondrial genomes to shed light on the evolutionary history of the two partners. Therefore, there is inevitably some repetition between chapters, as they share common themes. In the general introduction and discussion, I use the singular “I” as I am the sole author of these chapters. All other chapters are co-authored and therefore the plural “we” is used, including appendices belonging to these chapters. Part of chapter 2 has been published as: Bourguignon, T., Tang, Q., Ho, S.Y., Juna, F., Wang, Z., Arab, D.A., Cameron, S.L., Walker, J., Rentz, D., Evans, T.A. and Lo, N., 2018. Transoceanic dispersal and plate tectonics shaped global cockroach distributions: evidence from mitochondrial phylogenomics. Molecular Biology and Evolution, 35(4), pp.970-983. The chapter was reformatted to include additional data and analyses that I undertook towards this paper. My role was in the paper was to sequence samples, assemble mitochondrial genomes, perform phylogenetic analyses, and contribute to the writing of the manuscript.