Co-Circulation of Aedes Flavivirus, Culex Flavivirus, and Quang Binh
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

EN 14683 Report
Test Report NO.:BPF9JB4F41796710 Issued Date:2021-01-27 Page 1 of 3 Applicant: E. ERHARDT Y CIA,S.A. The following sample(s) was/were submitted and identified on behalf of the client as: Sample Name: Meltblown Fabric Sample Quantity/Amount: 1pcs Sample status description: Meltblown Fabric 2 meters long from 3 different rolls Sample Source: Send Sample Sample Received Date: 2021-01-21 Testing Period: 2021-01-21~2021-01-27 Test Item: Please refer to next page(s) Reference Methods: Please refer to next page(s) Test Result: Please refer to next page(s) Edited by: Checked by: Approved by: The page below is blank Hotline 400-819-5688 Pony Testing Group Shanghai Co.,Ltd. www.ponytest.com Company Address:2F.,No.7 Building, No.99, Wen Xiangdong Road,SongJiang District,Shanghai,China Tel:021-37895599 Test Address:No.5/6 Building,No.7/1F Building,No.7/3F Building, No.99, Wen Xiangdong Road, SongJiang District, Shanghai, China 2/3/4/6/F., Building 35, No.680, Guiping Road, Xuhui District, Shanghai, China Test Report NO.:BPF9JB4F41796710 Issued Date:2021-01-27 Page 2 of 3 Test Result Reference Methods: EN 14683:2019 +AC:2019 EN 14683:2019 Test Item +AC:2019 Limit Test Result Evaluation Type Ⅱ A B C 1-1 22.2 1-2 23.9 1-3 23.5 22.9 1-4 22.5 1-5 22.3 2-1 25.2 2-2 22.3 2-3 21.1 23.0 2-4 24.0 2-5 22.4 3-1 22.4 Differential pressure, 3-2 23.9 2 <40 Conform Pa/cm 3-3 20.0 22.2 3-4 22.0 3-5 22.8 4-1 21.9 4-2 22.7 4-3 23.0 22.7 4-4 24.5 4-5 21.5 5-1 23.7 5-2 22.9 5-3 21.7 23.0 5-4 24.1 5-5 22.5 No.1 98.7 No.2 >99.9 Bacterial filtration ≥98 No.3 >99.9 Conform efficiency(BFE),% No.4 99.9 No.5 99.7 Hotline 400-819-5688 Pony Testing Group Shanghai Co.,Ltd. -

2020 Shanghai Foreign Investment Guide Shanghai Foreign Shanghai Foreign Investment Guide Investment Guide
2020 SHANGHAI FOREIGN INVESTMENT GUIDE SHANGHAI FOREIGN SHANGHAI FOREIGN INVESTMENT GUIDE INVESTMENT GUIDE Contents Investment Chapter II Promotion 61 Highlighted Investment Areas 10 Institutions Preface 01 Overview of Investment Areas A Glimpse at Shanghai's Advantageous Industries Appendix 66 Chapter I A City Abundant in 03 Chapter III Investment Opportunities Districts and Functional 40 Enhancing Urban Capacities Zones for Investment and Core Functions Districts and Investment Influx of Foreign Investments into Highlights the Pioneer of China’s Opening-up Key Functional Zones Further Opening-up Measures in Support of Local Development SHANGHAI FOREIGN SHANGHAI FOREIGN 01 INVESTMENT GUIDE INVESTMENT GUIDE 02 Preface Situated on the east coast of China highest international standards Secondly, the openness of Shanghai Shanghai is becoming one of the most At the beginning of 2020, Shang- SHFTZ with a new area included; near the mouth of the Yangtze River, and best practices. As China’s most translates into a most desired invest- desired investment destinations for hai released the 3.0 version of its operating the SSE STAR Market with Shanghai is internationally known as important gateway to the world, ment destination in the world char- foreign investors. business environment reform plan its pilot registration-based IPO sys- a pioneer of China’s opening to the Shanghai has persistently functioned acterized by increasing vitality and Thirdly, the openness of Shanghai is – the Implementation Plan on Deep- tem; and promoting the integrated world for its inclusiveness, pursuit as a leader in the national opening- optimized business environment. shown in its pursuit of world-lead- ening the All-round Development of a development of the YRD region as of excellence, cultural diversity, and up initiative. -

Suburban Development in Shanghai: a Case of Songjiang
CARDIFF UNIVERSITY SCHOOL OF CITY AND REGIONAL PLANNING Suburban Development in Shanghai: A Case of Songjiang Jie Shen Supervisor: Professor Fulong Wu Thesis for the Degree of Doctor of Philosophy December 2011 ABSTRACT Since 2000, a new round of suburbanisation characterised by mixed-use clustered development has begun to unfold in China. This research aims to explore the dynamics of recent suburban growth in China and also provide an empirical case for enriching suburban theory. It is held that suburbanisation in China in its current form is by no means a spontaneous process, but results from capitalism’s creation of a new space to facilitate accumulation. Based on this view, the study examines the underlying forces of contemporary suburban growth with regard to three questions: what is the role of suburbanisation in China’s contemporary capital accumulation regime? How are the suburbs developed under coalitions of different actors? And how is suburban development shaped by demand-side actors? The study is founded on an intensive case study of Shanghai and one of its suburban districts, Songjiang. Both qualitative and quantitative research methods are used. Firsthand data from interviews and a questionnaire survey and a wide variety of secondary data were collected, providing a rich fund of knowledge for the research. While similar forms and functions to (post)-suburban settlements that have recently emerged in Western countries are found in Chinese suburbs, suburbanisation through new town development in China is a strategy of capital accumulation in response to a range of new conditions specific to China’s local context. New towns deal with the recentralisation of both fiscal and land development powers on the one hand, and accommodate the increasing housing demands of a diverse labour force on the other. -

The Oriental Pearl Radio & TV Tower 东方明珠 Getting in Redeem Your
The Oriental Pearl Radio & TV Tower 东方明珠 Getting In Redeem your pass for an admission ticket at the first ticket office, near No. 1 Gate. Hours Daily, 8:00 am-9:30 pm. Address No. 1 Lujiazui Century Ave Pudong New Area, Shanghai Public Transportation Take Metro Line 2 and get off at Lujiazui Station, get out from Exit 1 and walk to The Oriental Pearl Radio & TV Tower. Yu Garden (Yuyuan) 豫园 Getting In Please redeem your pass for an admission ticket at the Yuyuan Garden ticket office located on the north side of the Huxin Pavilion Jiuqu Bridge prior to entry. Hours Daily, 8:45 am-4:45 pm. Address No. 218 Anren St Huangpu District, Shanghai Public Transportation Take Metro Line 10 and get off at Yuyuan Station, then walk to Yu Garden. Shanghai World Financial Center Observatory 上海环球金融中心 Getting In Please redeem your pass for an admission ticket at the Global Finance Center F1 ticket window located at Lujiazui Century Ave. Hours Daily, 9:00 am-10:30 pm. Address B1 Ticketing Window, World Financial Center 100 Century Avenue Lujiazui, Pudong New Area, Shanghai Public Transportation Take Metro Line 2 and get off at Lujiazui Station, then walk to Shanghai World Financial Center. Shanghai Hop-On Hop-Off Sightseeing Bus Tour 观光巴士 Getting In You must first redeem your pass for a bus ticket at one of the following locations prior to boarding: Nanjing Road Station (New World City Stop): Opposite to New World City, No. 2-88 Nanjing West Road, Huangpu District, Shanghai Bund A Station (Sanyang Food Stop): Beside Sanyang Food, 367 East Zhongshan Road, Huangpu District, Shanghai (near Beijing East Road) Shiliupu Station (Pujiang Tour Terminal Stop): 531 Zhongshan East Second Road, Huangpu District, Shanghai Yuyuan Station (Yongan Road, Renmin Road): Xinkaihe Road, Renmin Road, next to the bus stop in front of the Bund soho. -
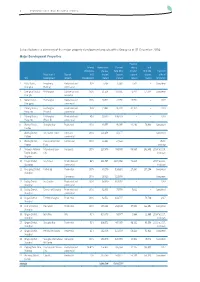
Major Development Properties
1 SHANGHAI INDUSTRIAL HOLDINGS LIMITED Set out below is a summary of the major property development projects of the Group as at 31 December 2016: Major Development Properties Pre-sold Interest Approximate Planned during Total attributable site area total GFA the year GFA sold Expected Projects of SI Type of to SI (square (square (square (square date of City Development property Development meters) meters) meters) meters) completion 1 Kaifu District, Fengsheng Residential and 90% 5,468 70,566 7,542 – Completed Changsha Building commercial 2 Chenghua District, Hi-Shanghai Commercial and 100% 61,506 254,885 75,441 151,644 Completed Chengdu residential 3 Beibei District, Hi-Shanghai Residential and 100% 30,845 74,935 20,092 – 2019 Chongqing commercial 4 Yuhang District, Hi-Shanghai Residential and 85% 74,864 230,484 81,104 – 2019 Hangzhou (Phase I) commercial 5 Yuhang District, Hi-Shanghai Residential and 85% 59,640 198,203 – – 2019 Hangzhou (Phase II) commercial 6 Wuxing District, Shanghai Bay Residential 100% 85,555 96,085 42,236 76,966 Completed Huzhou 7 Wuxing District, SIIC Garden Hotel Hotel and 100% 116,458 47,177 – – Completed Huzhou commercial 8 Wuxing District, Hurun Commercial Commercial 100% 13,661 27,322 – – Under Huzhou Plaza planning 9 Shilaoren National International Beer Composite 100% 227,675 783,500 58,387 262,459 2014 to 2018, Tourist Resort, City in phases Qingdao 10 Fengze District, Sea Palace Residential and 49% 381,795 1,670,032 71,225 – 2017 to 2021, Quanzhou commercial in phases 11 Changning District, United 88 Residential -

Shanghai Project Envision 2116 September 4, 2016 – July 30, 2017
Shanghai Project Envision 2116 September 4, 2016 – July 30, 2017 The inaugural edition of the Shanghai Project, Envision 2116, will launch the first of its two phases on September 4. Under the co-artistic directorship of Yongwoo Lee, Executive Director of Shanghai Himalayas Museum, and Hans Ulrich Obrist, Artistic Director of Serpentine Galleries London, the first edition of Shanghai Project focuses on public engagement through event-based programming, as well as exhibitions, public works, an open call platform, a pop–up library, community participation programs and international conferences. As one of the highlights of Phase 1, the Envision Pavilion, designed by Sou Fujimoto, will open to the public on September 4 at the Shanghai Himalayas Center. The structure, with a 670 square meter footprint, is a hybrid space composed of industrial scaffolding grids, glass and trees, combining notions of inside/outside, natural/man-made, organic/geometric and building/beyond-building. It serves as a symbolic center of the Shanghai Project. For the full duration of the Shanghai Project, the Envision Pavilion acts as a site for lectures, panels, workshops, seminars, performances and film screenings in the event space, as well as exhibitions in its gallery space and social gatherings in its café. In the gallery space, the audience will encounter Cildo Meireles‘s installation Ku Kka Ka Kka as well as Xu Bing‘s work from his series Background Story, and Jenova Chen‘s video game, Journey. Further, the pavilion will host Landversation, a series of performances developed by Otobong Nkanga and her team. Also in the event space, visitors will find the “Shanghai Project Pop-Up Library of the Future,” launched in collaboration with the renowned Shanghai Jifeng Bookstore. -

The Oriental Pearl Radio & TV Tower 东方明珠
The Oriental Pearl Radio & TV Tower 东方明珠 Hours: Daily, 9:00 am-9:30 pm. Address: No. 1 Century Ave Pudong New Area (Lujiazui), Shanghai Public Transportation Take Metro Line 2 and get off at Lujiazui Station, get out from Exit 1 and walk to The Oriental Pearl Radio & TV Tower. Getting In Redeem your pass for an admission ticket at the first ticket office, near No. 1 Gate: Shanghai World Financial Center Observatory 上海环球金融中心 Hours: Daily, 9:00 am-10:00 pm. Address: B1 Ticketing Window, World Financial Center 100 Century Avenue Lujiazui, Pudong New Area, Shanghai Public Transportation Take Metro Line 2 and get off at Lujiazui Station, then walk to Shanghai World Financial Center. Getting In Please redeem your pass for an admission ticket at B1 Ticketing Window, World Financial Center at Lujiazui Century Ave: Pujiang River Cruise Tour 黄浦江“清游江”游览船 Hours:Daily, 10:00 am-8:30 pm. Address:Shiliupu Cruise Terminal,No. 481 Zongshan Rd,Huangpu District, Shanghai Public Transportation Bus: Take the bus #33, 55, 65, 305, 868, 910, 926 or 928 and get off at the Xinkaihe Road-Bus Stop of Zhongshan East Second Road, then walk to No. 481, Zhongshan East Second Road, Huangpu District. Getting In Redeem your pass for an admission ticket at the Shiliu Pu Pier, Huangpu River Tour ticket window at 481 Zhongshan 2nd Rd: Yu Garden (Yuyuan) 豫园 Hours: Daily, 8:45 am-4:45 pm. Address: No. 218 Anren St Huangpu District, Shanghai Public Transportation Take Metro Line 10 and get off at Yuyuan Station, then walk to Yu Garden. -
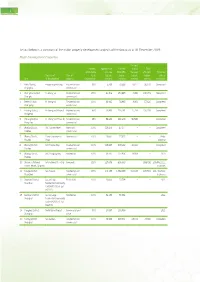
Set out Below Is a Summary of the Major Property Development Projects of the Group As at 31 December 2019: Major Development
1 Set out below is a summary of the major property development projects of the Group as at 31 December 2019: Major Development Properties Pre-sold Interest Approximate Planned during Total attributable site area total GFA the year GFA sold Expected Projects of Type of to SI (square (square (square (square date of City SI Development property Development meters) meters) meters) meters) completion 1 Kaifu District, Fengsheng Building Residential and 90% 5,468 70,566 6,627 30,870 Completed Changsha commercial 2 Chenghua District, Hi-Shanghai Residential and 100% 61,506 254,885 4,996 190,153 Completed Chengdu commercial 3 Beibei District, Hi-Shanghai Residential and 100% 30,845 74,935 3,301 57,626 Completed Chongqing commercial 4 Yuhang District, Hi-Shanghai (Phase I) Residential and 85% 74,864 230,484 27,758 150,289 Completed Hangzhou commercial 5 Yuhang District, Hi-Shanghai (Phase II) Residential and 85% 59,640 198,203 56,539 – Completed Hangzhou commercial 6 Wuxing District, SIIC Garden Hotel Hotel and 100% 116,458 47,177 – – Completed Huzhou commercial 7 Wuxing District, Hurun Commercial Commercial 100% 13,661 27,322 – – Under Huzhou Plaza planning 8 Wuxing District, SIIC Tianlan Bay Residential and 100% 115,647 193,292 26,042 – Completed Huzhou commercial 9 Wuxing District, SIIC Yungjing Bay Residential 100% 68,471 207,906 28,953 – 2020 Huzhou 10 Shilaoren National International Beer City Composite 100% 227,675 806,339 – 333,798 2014 to 2022, Tourist Resort, Qingdao in phases 11 Fengze District, Sea Palace Residential and 100% 170,133 -

Real Estate and Construction-210204-EN
Real Estate and Construction Beijing Guangzhou Hong Kong Shanghai Shenzhen 27/F, North Tower 17/F, International Finance 26/F, One Exchange Square 24/F, HKRI Centre Two, 17/F, Tower One, Kerry Plaza Beijing Kerry Centre Place, 8 Huaxia Road, 8 Connaught Place, Central HKRI Taikoo Hui 1 Zhong Xin Si Road 1 Guanghua Road Zhujiang New Town Hong Kong 288 Shi Men Yi Road Futian District Chaoyang District Guangzhou 510623, China Shanghai 200041, China Shenzhen 518048, China Beijing 100020, China Tel: +86 10 5769 5600 Tel: +86 20 3225 3888 Tel: +852 3976 8888 Tel: +86 21 2208 1166 Tel: +86 755 8159 3999 Fax:+86 10 5769 5788 Fax:+86 20 3225 3899 Fax:+852 2110 4285 Fax:+86 21 5298 5599 Fax:+86 755 8159 3900 www.fangdalaw.com Real Estate and Construction 01 Real Estate and Construction Practice Fangda’s Real Estate and Construction Practice Team excels at providing one-stop and full-scale services to leading real estate market players, including real estate developers, real estate private equity sponsors, institutional investors, financial institutions and other corporations and individuals in complicated domestic and cross-border real estate transactions. Our lawyers are frequently appointed as arbitrators in real estate and construction-related disputes. Our strength in this area includes a combination of understanding the traditions of the Chinese market and practice and our abundant experience in serving international clients, complemented by services for non-contentious and contentious cases. We are one of the few firms based in China that can provide integrated real estate and construction related legal services, and present practical advice based on our knowledge and experience in the local market. -

Development of High-Speed Rail in the People's Republic of China
A Service of Leibniz-Informationszentrum econstor Wirtschaft Leibniz Information Centre Make Your Publications Visible. zbw for Economics Haixiao, Pan; Ya, Gao Working Paper Development of high-speed rail in the People's Republic of China ADBI Working Paper Series, No. 959 Provided in Cooperation with: Asian Development Bank Institute (ADBI), Tokyo Suggested Citation: Haixiao, Pan; Ya, Gao (2019) : Development of high-speed rail in the People's Republic of China, ADBI Working Paper Series, No. 959, Asian Development Bank Institute (ADBI), Tokyo This Version is available at: http://hdl.handle.net/10419/222726 Standard-Nutzungsbedingungen: Terms of use: Die Dokumente auf EconStor dürfen zu eigenen wissenschaftlichen Documents in EconStor may be saved and copied for your Zwecken und zum Privatgebrauch gespeichert und kopiert werden. personal and scholarly purposes. Sie dürfen die Dokumente nicht für öffentliche oder kommerzielle You are not to copy documents for public or commercial Zwecke vervielfältigen, öffentlich ausstellen, öffentlich zugänglich purposes, to exhibit the documents publicly, to make them machen, vertreiben oder anderweitig nutzen. publicly available on the internet, or to distribute or otherwise use the documents in public. Sofern die Verfasser die Dokumente unter Open-Content-Lizenzen (insbesondere CC-Lizenzen) zur Verfügung gestellt haben sollten, If the documents have been made available under an Open gelten abweichend von diesen Nutzungsbedingungen die in der dort Content Licence (especially Creative Commons Licences), you genannten Lizenz gewährten Nutzungsrechte. may exercise further usage rights as specified in the indicated licence. https://creativecommons.org/licenses/by-nc-nd/3.0/igo/ www.econstor.eu ADBI Working Paper Series DEVELOPMENT OF HIGH-SPEED RAIL IN THE PEOPLE’S REPUBLIC OF CHINA Pan Haixiao and Gao Ya No. -
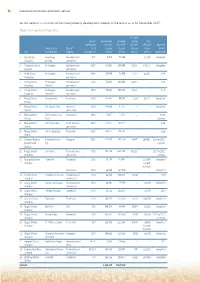
1 Set out Below Is a Summary of the Major Property Development
1 SHANGHAI INDUSTRIAL HOLDINGS LIMITED Set out below is a summary of the major property development projects of the Group as at 31 December 2017: Major Development Properties Pre-sold Interest Approximate Planned during Total attributable site area total GFA the year GFA sold Expected Projects of SI Type of to SI (square (square (square (square date of City Development property Development meters) meters) meters) meters) completion 1 Kaifu District, Fengsheng Residential and 90% 5,468 70,144 – 12,538 Completed Changsha Building commercial 2 Chenghua District, Hi-Shanghai Residential and 100% 61,506 254,885 14,355 176,157 Completed Chengdu commercial 3 Beibei District, Hi-Shanghai Residential and 100% 30,843 71,459 27,707 44,682 2019 Chongqing commercial 4 Yuhang District, Hi-Shanghai Residential and 85% 74,864 230,484 55,910 – 2018 Hangzhou (Phase I) commercial 5 Yuhang District, Hi-Shanghai Residential and 100% 59,640 198,203 23,001 – 2019 Hangzhou (Phase II) commercial 6 Wuxing District, Shanghai Bay Residential 100% 85,555 96,085 2,559 86,122 Completed Huzhou 7 Wuxing District, SIIC Garden Hotel Hotel and 100% 116,458 47,177 – – Completed Huzhou commercial 8 Wuxing District, Hurun Commercial Commercial 100% 13,661 27,322 – – Under Huzhou Plaza planning 9 Wuxing District, SIIC Tianlan Bay Residential and 100% 115,647 180,211 – – 2019 Huzhou commercial 10 Wuxing District, SIIC Yungjing Bay Residential 100% 68,471 203,087 – – 2020 Huzhou 11 Shilaoren National International Beer Composite 100% 227,675 787,000 14,547 295,661 2014 to 2018, -

Quarterly Newsletter of GEF China Sustainable Cities Integrated Approach Pilot Project
Quarterly Newsletter of GEF China Sustainable Cities Integrated Approach Pilot Project issue 8 June 2020 Project Progress (As of June 15, 2020) GEBJ-2: The evaluation for the technical proposal was completed on May 19, 2020. Ministry of Housing and Urban-Rural The bid opening for the financial proposal and Development of P.R.C. contract negotiation were held on June 11, 2020. The PMO intends to partially adjust the The contract of the National TOD Platform tasks in the TOR. A written request of specific was officially signed on April 20, 2020. The changes will be submitted to the World Bank Project Management Office (PMO) held the task team by the end of June 2020. kick-off meeting for the hired consultant to GEBJ-3: Request of Expression of present the inception report and work plans on Interest (REOI) was posted on April 28, May 29, 2020. The inception report and work 2020. The shortlist of qualified bidders was plans were reviewed by a panel of experts on evaluated on June 10, 2020. RFP is currently June 15. It will be finalized and submitted to under preparation and will be sent to the the World Bank task team by the end of June qualified bidders by the end of June 2020. 2020. Tianjin GETJ-1: The first draft for Task 5: The Contextualized TOD Guidebook and Toolkit for Tianjin was completed at the beginning of May 2020. The final draft will be completed at the end of June 2020. The disbursement of grant submitted to the World Bank task team after the evaluation for the final draft is completed.