Resistance to the CHK1 Inhibitor Prexasertib Involves Functionally Distinct CHK1 Activities in BRCA Wild-Type Ovarian Cancer
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Telomeres.Pdf
Telomeres Secondary article Elizabeth H Blackburn, University of California, San Francisco, California, USA Article Contents . Introduction Telomeres are specialized DNA–protein structures that occur at the ends of eukaryotic . The Replication Paradox chromosomes. A special ribonucleoprotein enzyme called telomerase is required for the . Structure of Telomeres synthesis and maintenance of telomeric DNA. Synthesis of Telomeric DNA by Telomerase . Functions of Telomeres Introduction . Telomere Homeostasis . Alternatives to Telomerase-generated Telomeric DNA Telomeres are the specialized chromosomal DNA–protein . Evolution of Telomeres and Telomerase structures that comprise the terminal regions of eukaryotic chromosomes. As discovered through studies of maize and somes. One critical part of this protective function is to fruitfly chromosomes in the 1930s, they are required to provide a means by which the linear chromosomal DNA protect and stabilize the genetic material carried by can be replicated completely, without the loss of terminal eukaryotic chromosomes. Telomeres are dynamic struc- DNA nucleotides from the 5’ end of each strand of this tures, with their terminal DNA being constantly built up DNA. This is necessary to prevent progressive loss of and degraded as dividing cells replicate their chromo- terminal DNA sequences in successive cycles of chromo- somes. One strand of the telomeric DNA is synthesized by somal replication. a specialized ribonucleoprotein reverse transcriptase called telomerase. Telomerase is required for both -

Ailanthone Inhibits Non-Small Cell Lung Cancer Cell Growth Through Repressing DNA Replication Via Downregulating RPA1
FULL PAPER British Journal of Cancer (2017) 117, 1621–1630 | doi: 10.1038/bjc.2017.319 Keywords: ailanthone; non-small cell lung cancer; DNA replication; RPA1; Chinese medicine Ailanthone inhibits non-small cell lung cancer cell growth through repressing DNA replication via downregulating RPA1 Zhongya Ni1, Chao Yao1, Xiaowen Zhu1, Chenyuan Gong1, Zihang Xu2, Lixin Wang3, Suyun Li4, Chunpu Zou2 and Shiguo Zhu*,1,3 1Laboratory of Integrative Medicine, School of Basic Medical Sciences, Shanghai University of Traditional Chinese Medicine, 1200 Cai Lun Rd, Shanghai 201203, PR China; 2Department of Internal Classic of Medicine, School of Basic Medical Sciences, Shanghai University of Traditional Chinese Medicine, 1200 Cai Lun Rd, Shanghai 201203, PR China; 3Department of Immunology and Pathogenic Biology, School of Basic Medical Sciences, Shanghai University of Traditional Chinese Medicine, 1200 Cai Lun Rd, Shanghai 201203, PR China and 4Department of Pathology, School of Basic Medical Sciences, Shanghai University of Traditional Chinese Medicine, 1200 Cai Lun Rd, Shanghai 201203, PR China Background: The identification of bioactive compounds from Chinese medicine plays a crucial role in the development of novel reagents against non-small cell lung cancer (NSCLC). Methods: High throughput screening assay and analyses of cell growth, cell cycle, apoptosis, cDNA microarray, BrdU incorporation and gene expression were performed. Results: Ailanthone (Aila) suppressed NSCLC cell growth and colony formation in vitro and inhibited NSCLC tumour growth in subcutaneously xenografted and orthotopic lung tumour models, leading to prolonged survival of tumour-bearing mice. Moreover, Aila induced cell cycle arrest in a dose-independent manner but did not induce apoptosis in all NSCLC cells. -

Iron Depletion Reduces Abce1 Transcripts While Inducing The
Preprints (www.preprints.org) | NOT PEER-REVIEWED | Posted: 22 October 2019 doi:10.20944/preprints201910.0252.v1 1 Research Article 2 Iron depletion Reduces Abce1 Transcripts While 3 Inducing the Mitophagy Factors Pink1 and Parkin 4 Jana Key 1,2, Nesli Ece Sen 1, Aleksandar Arsovic 1, Stella Krämer 1, Robert Hülse 1, Suzana 5 Gispert-Sanchez 1 and Georg Auburger 1,* 6 1 Experimental Neurology, Goethe University Medical School, 60590 Frankfurt am Main; 7 2 Faculty of Biosciences, Goethe-University Frankfurt am Main, Germany 8 * Correspondence: [email protected] 9 10 Abstract: Lifespan extension was recently achieved in Caenorhabditis elegans nematodes by 11 mitochondrial stress and mitophagy, triggered via iron depletion. Conversely in man, deficient 12 mitophagy due to Pink1/Parkin mutations triggers iron accumulation in patient brain and limits 13 survival. We now aimed to identify murine fibroblast factors, which adapt their mRNA expression 14 to acute iron manipulation, relate to mitochondrial dysfunction and may influence survival. After 15 iron depletion, expression of the plasma membrane receptor Tfrc with its activator Ireb2, the 16 mitochondrial membrane transporter Abcb10, the heme-release factor Pgrmc1, the heme- 17 degradation enzyme Hmox1, the heme-binding cholesterol metabolizer Cyp46a1, as well as the 18 mitophagy regulators Pink1 and Parkin showed a negative correlation to iron levels. After iron 19 overload, these factors did not change expression. Conversely, a positive correlation of mRNA levels 20 with both conditions of iron availability was observed for the endosomal factors Slc11a2 and Steap2, 21 as well as for the iron-sulfur-cluster (ISC)-containing factors Ppat, Bdh2 and Nthl1. -
Supplementary Text and Figures.Pdf
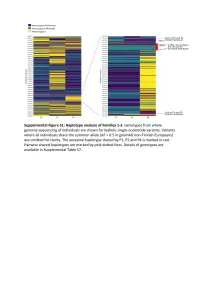
Supplemental Figure S2. PRIM1 immunoblots (20-75 kDa). (A) Corresponds to Figure 2D, with additional bands seen in P2 at 75 kDa and 25 kDa. (B) Independent experiment with cell lysates from C and P2 in which additional bands at 25 kDa not evident. (C) Corresponds to Figure 2D. No additional band visible at 25 kDa. (D) Validation of monoclonal antibody (8G10) with siRNA to PRIM1 in HeLa cells. siLUC, luciferase negative control. siPRIM1, siRNA targeting PRIM1 transcript. Vinculin used as a loading control. Supplemental Figure S3: Predicted destabilisation of PRIM1 by the C301R substitution. (A) Cysteine 301 lies in a buried hydrophobic region in PRIM1. DNA primase dimer crystal structure (PDB: 4BPU). PRIM1 residues shaded according to solvent accessibility. Cysteine 301, substituted to arginine in P5, lies in a buried hydrophobic region. (B) Cys301 is evolutionarily conserved in vertebrates. Neither arginine or other large or charged amino acids are observed at this position in other species. (C) The C301R substitution observed in P5 is predicted to lead to destabilisation of all available PRIM1 crystal structures. In contrast, substitution with leucine or threonine, as observed in some orthologous proteins, is not predicted to lead to destabilisation. Data points, predicted changes in free-energy (ΔΔG) from FoldX plotted for each of 14 available crystal structures. (D) Immunoblotting of RPE1 cells transfected with dual reporter constructs (as described in Figure 3B and Materials and Methods) confirms production of PRIM1-GFP and FLAG-SR at expected molecular weights and shows reduced protein levels for the C301R variant. n=2 experiments shown. C301R protein levels relative to WT after normalization to actin loading control indicated underneath the blot. -
Synthetic Lethality Between DNA Polymerase Epsilon and RTEL1 in Metazoan DNA Replication
Article Synthetic Lethality between DNA Polymerase Epsilon and RTEL1 in Metazoan DNA Replication Graphical Abstract Authors Roberto Bellelli, Jillian Youds, Valerie Borel, Jennifer Svendsen, Visnja Pavicic-Kaltenbrunner, Simon J. Boulton Correspondence [email protected] In Brief Bellelli et al. report that RTEL1 deficiency is synthetic lethal with the loss of pole-4 in C. elegans/hypomorphy of Pol epsilon. An analysis of replication dynamics in Rtel1À/À Pole4À/À mouse cells revealed a combination of dysfunctional fork progression and defective origin activation, which cooperatively drive incomplete genomic replication and genetic instability. Highlights d rtel-1 is synthetic lethal with the loss of DNA polymerase epsilon in C. elegans d rtel-1; pole-4 double mutants accumulate Rad51 and RPA foci and fail to replicate d Impaired DNA replication and genome instability in Rtel1 Pole4 knockout mouse cells d Rtel1 Pole4 double knockout mouse cells exhibit fork asymmetry and defective origin activation Bellelli et al., 2020, Cell Reports 31, 107675 May 26, 2020 ª 2020 The Author(s). https://doi.org/10.1016/j.celrep.2020.107675 ll ll OPEN ACCESS Article Synthetic Lethality between DNA Polymerase Epsilon and RTEL1 in Metazoan DNA Replication Roberto Bellelli,1,2,3 Jillian Youds,1,2 Valerie Borel,1 Jennifer Svendsen,1 Visnja Pavicic-Kaltenbrunner,1 and Simon J. Boulton1,4,* 1The Francis Crick Institute, 1 Midland Road, NW1 1AT London, UK 2These authors contributed equally 3Present address: Center for Cancer Cell and Molecular Biology, Barts Cancer Institute, Queen Mary University of London, Charterhouse Square, Barbican, EC1M 6BE London, UK 4Lead Contact *Correspondence: [email protected] https://doi.org/10.1016/j.celrep.2020.107675 SUMMARY Genome stability requires coordination of DNA replication origin activation and replication fork progression. -

Cell Growth-Regulated Expression of Mammalian MCM5 and MCM6 Genes Mediated by the Transcription Factor E2F
Oncogene (1999) 18, 2299 ± 2309 ã 1999 Stockton Press All rights reserved 0950 ± 9232/99 $12.00 http://www.stockton-press.co.uk/onc Cell growth-regulated expression of mammalian MCM5 and MCM6 genes mediated by the transcription factor E2F Kiyoshi Ohtani1, Ritsuko Iwanaga1, Masataka Nakamura*,1, Masa-aki Ikeda2, Norikazu Yabuta3, Hiromichi Tsuruga3 and Hiroshi Nojima3 1Human Gene Sciences Center, Tokyo Medical and Dental University, Tokyo 113-8510, Japan 2Department of Developmental Biology, Graduate School of Dentistry, Tokyo Medical and Dental University, Tokyo 113-8549, Japan; 3Department of Molecular Genetics, Research Institute for Microbial Diseases, Osaka University, Suita 565-0871, Japan Initiation of DNA replication requires the function of family (MCM2-7) that have been identi®ed in yeast, MCM gene products, which participate in ensuring that Xenopus, and human. Mcm proteins seem to regulate DNA replication occurs only once in the cell cycle. the initiation at the replication origin where the loading Expression of all mammalian genes of the MCM family of the proteins onto the origin recognition complex is induced by growth stimulation, unlike yeast, and the (ORC) is regulated by Cdc6 and cyclin-dependent mRNA levels peak at G1/S boundary. In this study, we kinases (Donovan et al., 1997; Tanaka et al., 1997). examined the transcriptional activities of isolated human However, the mechanism(s) by which Mcm proteins MCM gene promoters. Human MCM5 and MCM6 control the initiation of DNA replication remains promoters with mutation in the E2F sites failed in unclear. promoter regulation following serum stimulation and Xenopus Mcm proteins seem to be able to access exogenous E2F expression. -

Replication Stress: an Achilles' Heel of Glioma Cancer Stem–Like Cells Meredith A
Published OnlineFirst November 29, 2018; DOI: 10.1158/0008-5472.CAN-18-2439 Cancer Review Research Replication Stress: An Achilles' Heel of Glioma Cancer Stem–like Cells Meredith A. Morgan1, and Christine E. Canman2 Abstract Glioblastoma (GBM) is a highly aggressive form of cancer that Carruthers and colleagues investigated DNA replication stress as is resistant to standard therapy with concurrent radiation and an underlying mechanism responsible for upregulation of the temozolomide, two agents that work by inducing DNA damage. DDR and hence the radiation resistance of glioma stem–like An underlying causeof this resistance may be a subpopulation of cells. Furthermore, the authors explore the efficacy of combined cancer stem–like cells that display a heightened DNA damage ataxia telangiectasia and Rad3-related kinase and PARP inhibi- response (DDR). Although this DDR represents an attractive tors as a strategy to leverage these mechanisms and overcome therapeutic target for overcoming the resistance of GBMs to radiation resistance. Cancer Res; 78(24); 6713–6. Ó2018 AACR. radiotherapy, until now, the cause of this DDR upregulation has See related article by Carruthers and colleagues, Cancer Res; not been understood. In a previous issue of Cancer Research, 78(17); 5060–71. The cancer stem cell theory states that a small subpopulation of of replication factories compared with non-GSC populations tumor cells possess unique self-renewal properties that are capa- (6). These observations pointed to elevated levels of RS as ble of seeding new tumors and are a source of regrowth following causative of DDR activation in untreated GSCs, a hypothesis therapy (1). Glioma stem–like cells (GSC) are defined as CD133- supported by the high levels of RS in glioblastoma (GBM; positive cells that can initiate new tumors in mice (2). -

POLE2 Knockdown Reduce Tumorigenesis in Esophageal Squamous Cells Yongjun Zhu, Gang Chen, Yang Song, Zhiming Chen* and Xiaofeng Chen*
Zhu et al. Cancer Cell Int (2020) 20:388 https://doi.org/10.1186/s12935-020-01477-4 Cancer Cell International PRIMARY RESEARCH Open Access POLE2 knockdown reduce tumorigenesis in esophageal squamous cells Yongjun Zhu, Gang Chen, Yang Song, Zhiming Chen* and Xiaofeng Chen* Abstract Background: Esophageal squamous cell carcinoma (ESCC) is one of the most frequent malignant tumors originated from digestive system around the world and the treatment was limited by the unclear mechanism. DNA polymerase epsilon 2, accessory subunit (POLE2) is involved in DNA replication, repair, and cell cycle control, whose association with ESCC is still not clear. Methods: In this study, the expression level of POLE2 in ESCC tissues was detected by IHC. The POLE2 knockdown cell line was constructed, identifed by qPCR and western blot and used for detecting cellular functions and con- structing xenotransplantation mice model. MTT Assay, colony formation assay, fow cytometry, wound-healing assay and Transwell assay were used to detected cell proliferation, apoptosis and migration. Results: We frstly identifed that the expression of POLE2 was overexpressed in ESCC. Moreover, the high expres- sion of POLE2 can predict the tumor deterioration and poor prognosis of ESCC patients. Additionally, downregulation of POLE2 was involved in ESCC progression by promoting proliferation, migration, and inhibiting apoptosis in vitro. In vivo studies proved that POLE2 was positively correlated with ESCC tumor formation, which was consistent with the results in vitro. We also illuminated that POLE2 knockdown upregulated pro-apoptotic proteins (Bax, Caspase3, CD40L, FasL, IGFBP-5 and P21) and downregulated anti-apoptotic proteins (CLAP-2, IGF-I and sTNF-R2). -

Bioinformatics-Based Screening of Key Genes for Transformation of Liver
Jiang et al. J Transl Med (2020) 18:40 https://doi.org/10.1186/s12967-020-02229-8 Journal of Translational Medicine RESEARCH Open Access Bioinformatics-based screening of key genes for transformation of liver cirrhosis to hepatocellular carcinoma Chen Hao Jiang1,2, Xin Yuan1,2, Jiang Fen Li1,2, Yu Fang Xie1,2, An Zhi Zhang1,2, Xue Li Wang1,2, Lan Yang1,2, Chun Xia Liu1,2, Wei Hua Liang1,2, Li Juan Pang1,2, Hong Zou1,2, Xiao Bin Cui1,2, Xi Hua Shen1,2, Yan Qi1,2, Jin Fang Jiang1,2, Wen Yi Gu4, Feng Li1,2,3 and Jian Ming Hu1,2* Abstract Background: Hepatocellular carcinoma (HCC) is the most common type of liver tumour, and is closely related to liver cirrhosis. Previous studies have focussed on the pathogenesis of liver cirrhosis developing into HCC, but the molecular mechanism remains unclear. The aims of the present study were to identify key genes related to the transformation of cirrhosis into HCC, and explore the associated molecular mechanisms. Methods: GSE89377, GSE17548, GSE63898 and GSE54236 mRNA microarray datasets from Gene Expression Omni- bus (GEO) were analysed to obtain diferentially expressed genes (DEGs) between HCC and liver cirrhosis tissues, and network analysis of protein–protein interactions (PPIs) was carried out. String and Cytoscape were used to analyse modules and identify hub genes, Kaplan–Meier Plotter and Oncomine databases were used to explore relationships between hub genes and disease occurrence, development and prognosis of HCC, and the molecular mechanism of the main hub gene was probed using Kyoto Encyclopedia of Genes and Genomes(KEGG) pathway analysis. -

Supplementary Table S1. Correlation Between the Mutant P53-Interacting Partners and PTTG3P, PTTG1 and PTTG2, Based on Data from Starbase V3.0 Database
Supplementary Table S1. Correlation between the mutant p53-interacting partners and PTTG3P, PTTG1 and PTTG2, based on data from StarBase v3.0 database. PTTG3P PTTG1 PTTG2 Gene ID Coefficient-R p-value Coefficient-R p-value Coefficient-R p-value NF-YA ENSG00000001167 −0.077 8.59e-2 −0.210 2.09e-6 −0.122 6.23e-3 NF-YB ENSG00000120837 0.176 7.12e-5 0.227 2.82e-7 0.094 3.59e-2 NF-YC ENSG00000066136 0.124 5.45e-3 0.124 5.40e-3 0.051 2.51e-1 Sp1 ENSG00000185591 −0.014 7.50e-1 −0.201 5.82e-6 −0.072 1.07e-1 Ets-1 ENSG00000134954 −0.096 3.14e-2 −0.257 4.83e-9 0.034 4.46e-1 VDR ENSG00000111424 −0.091 4.10e-2 −0.216 1.03e-6 0.014 7.48e-1 SREBP-2 ENSG00000198911 −0.064 1.53e-1 −0.147 9.27e-4 −0.073 1.01e-1 TopBP1 ENSG00000163781 0.067 1.36e-1 0.051 2.57e-1 −0.020 6.57e-1 Pin1 ENSG00000127445 0.250 1.40e-8 0.571 9.56e-45 0.187 2.52e-5 MRE11 ENSG00000020922 0.063 1.56e-1 −0.007 8.81e-1 −0.024 5.93e-1 PML ENSG00000140464 0.072 1.05e-1 0.217 9.36e-7 0.166 1.85e-4 p63 ENSG00000073282 −0.120 7.04e-3 −0.283 1.08e-10 −0.198 7.71e-6 p73 ENSG00000078900 0.104 2.03e-2 0.258 4.67e-9 0.097 3.02e-2 Supplementary Table S2. -

Gene Expression Profiling Analysis Contributes to Understanding the Association Between Non-Syndromic Cleft Lip and Palate, and Cancer
2110 MOLECULAR MEDICINE REPORTS 13: 2110-2116, 2016 Gene expression profiling analysis contributes to understanding the association between non-syndromic cleft lip and palate, and cancer HONGYI WANG, TAO QIU, JIE SHI, JIULONG LIANG, YANG WANG, LIANGLIANG QUAN, YU ZHANG, QIAN ZHANG and KAI TAO Department of Plastic Surgery, General Hospital of Shenyang Military Area Command, PLA, Shenyang, Liaoning 110016, P.R. China Received March 10, 2015; Accepted December 18, 2015 DOI: 10.3892/mmr.2016.4802 Abstract. The present study aimed to investigate the for NSCL/P were implicated predominantly in the TGF-β molecular mechanisms underlying non-syndromic cleft lip, signaling pathway, the cell cycle and in viral carcinogenesis. with or without cleft palate (NSCL/P), and the association The TP53, CDK1, SMAD3, PIK3R1 and CASP3 genes were between this disease and cancer. The GSE42589 data set found to be associated, not only with NSCL/P, but also with was downloaded from the Gene Expression Omnibus data- cancer. These results may contribute to a better understanding base, and contained seven dental pulp stem cell samples of the molecular mechanisms of NSCL/P. from children with NSCL/P in the exfoliation period, and six controls. Differentially expressed genes (DEGs) were Introduction screened using the RankProd method, and their potential functions were revealed by pathway enrichment analysis and Non-syndromic cleft lip, with or without cleft palate (NSCL/P) construction of a pathway interaction network. Subsequently, is one of the most common types of congenital defect and cancer genes were obtained from six cancer databases, and affects 3.4-22.9/10,000 individuals worldwide (1). -

Polymerase Δ Deficiency Causes Syndromic Immunodeficiency with Replicative Stress
Polymerase δ deficiency causes syndromic immunodeficiency with replicative stress Cecilia Domínguez Conde, … , Mirjam van der Burg, Kaan Boztug J Clin Invest. 2019. https://doi.org/10.1172/JCI128903. Research Article Genetics Immunology Graphical abstract Find the latest version: https://jci.me/128903/pdf The Journal of Clinical Investigation RESEARCH ARTICLE Polymerase δ deficiency causes syndromic immunodeficiency with replicative stress Cecilia Domínguez Conde,1,2 Özlem Yüce Petronczki,1,2,3 Safa Baris,4,5 Katharina L. Willmann,1,2 Enrico Girardi,2 Elisabeth Salzer,1,2,3,6 Stefan Weitzer,7 Rico Chandra Ardy,1,2,3 Ana Krolo,1,2,3 Hanna Ijspeert,8 Ayca Kiykim,4,5 Elif Karakoc-Aydiner,4,5 Elisabeth Förster-Waldl,9 Leo Kager,6 Winfried F. Pickl,10 Giulio Superti-Furga,2,11 Javier Martínez,7 Joanna I. Loizou,2 Ahmet Ozen,4,5 Mirjam van der Burg,8 and Kaan Boztug1,2,3,6 1Ludwig Boltzmann Institute for Rare and Undiagnosed Diseases, 2CeMM Research Center for Molecular Medicine of the Austrian Academy of Sciences, and 3St. Anna Children’s Cancer Research Institute (CCRI), Vienna, Austria. 4Pediatric Allergy and Immunology, Marmara University, Faculty of Medicine, Istanbul, Turkey. 5Jeffrey Modell Diagnostic Center for Primary Immunodeficiency Diseases, Marmara University, Istanbul, Turkey. 6St. Anna Children’s Hospital, Department of Pediatrics and Adolescent Medicine, Vienna, Austria. 7Center for Medical Biochemistry, Medical University of Vienna, Vienna, Austria. 8Department of Pediatrics, Laboratory for Immunology, Leiden University Medical Centre, Leiden, Netherlands. 9Department of Neonatology, Pediatric Intensive Care and Neuropediatrics, Department of Pediatrics and Adolescent Medicine, 10Institute of Immunology, Center for Pathophysiology, Infectiology and Immunology, and 11Center for Physiology and Pharmacology, Medical University of Vienna, Vienna, Austria.