Mutation Rates in Seeds and Seed-Banking Influence Substitution Rates Across the Angiosperm Phylogeny
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Tesis. Síndromes De Polinización En
Dr. Luis Giménez Benavides, Profesor Contratado Doctor del Departamento de Biología y Geología, Física y Química Inorgánica de la Universidad Rey Juan Carlos, CERTIFICA Que los trabajos de investigación desarrollados en la memoria de tesis doctoral, “Síndromes de polinización en Silene. Evolución de las interacciones polinizador-depredador con Hadena” son aptos para ser presentados por el Ldo. Samuel Prieto Benítez ante el tribunal que en su día se consigne, para aspirar al Grado de Doctor en el Programa de Doctorado de Conservación de Recursos Naturales por la Universidad Rey Juan Carlos de Madrid. V°B° Director de Tesis Dr. Luis Giménez Benavides TESIS DOCTORAL Síndromes de polinización en Silene. Evolución de las interacciones polinizador- depredador con Hadena. Samuel Prieto Benítez Dirigida por: Luis Giménez Benavides Departamento de Biología y Geología, Física y Química Inorgánica Universidad Rey Juan Carlos Mayo 2015 A mi familia y a Sofía, gracias por el apoyo y el cariño que me dais. ÍNDICE RESUMEN Antecedentes 11 Objetivos 19 Metodología 20 Conclusiones 25 Referencias 27 Lista de manuscritos 33 CAPÍTULOS/CHAPTERS Capítulo 1/Chapter 1 35 Revisión y actualización del estado de conocimiento de las relaciones polinización- depredación entre Caryophyllaceae y Hadena (Noctuidae). Capítulo 2/Chapter 2 65 Diel Variation in Flower Scent Reveals Poor Consistency of Diurnal and Nocturnal Pollination Syndromes in Sileneae. Capítulo 3/Chapter 3 113 Floral scent evolution in Silene: a multivariate phylogenetic analysis. Capítulo 4/Chapter 4 145 Flower circadian rhythm restricts/constraints pollination generalization and prevents the escape from a pollinator-seed predating specialist in Silene. Capítulo 5/Chapter 5 173 Spatio-temporal variation in the interaction outcome between a nursery pollinator and its host plant when other other pollinators, fruit predators and nectar robbers are present. -

"National List of Vascular Plant Species That Occur in Wetlands: 1996 National Summary."
Intro 1996 National List of Vascular Plant Species That Occur in Wetlands The Fish and Wildlife Service has prepared a National List of Vascular Plant Species That Occur in Wetlands: 1996 National Summary (1996 National List). The 1996 National List is a draft revision of the National List of Plant Species That Occur in Wetlands: 1988 National Summary (Reed 1988) (1988 National List). The 1996 National List is provided to encourage additional public review and comments on the draft regional wetland indicator assignments. The 1996 National List reflects a significant amount of new information that has become available since 1988 on the wetland affinity of vascular plants. This new information has resulted from the extensive use of the 1988 National List in the field by individuals involved in wetland and other resource inventories, wetland identification and delineation, and wetland research. Interim Regional Interagency Review Panel (Regional Panel) changes in indicator status as well as additions and deletions to the 1988 National List were documented in Regional supplements. The National List was originally developed as an appendix to the Classification of Wetlands and Deepwater Habitats of the United States (Cowardin et al.1979) to aid in the consistent application of this classification system for wetlands in the field.. The 1996 National List also was developed to aid in determining the presence of hydrophytic vegetation in the Clean Water Act Section 404 wetland regulatory program and in the implementation of the swampbuster provisions of the Food Security Act. While not required by law or regulation, the Fish and Wildlife Service is making the 1996 National List available for review and comment. -

Complete Sequence and Comparative Analysis of the Chloroplast Genome of Coconut Palm (Cocos Nucifera)
Complete Sequence and Comparative Analysis of the Chloroplast Genome of Coconut Palm (Cocos nucifera) Ya-Yi Huang, Antonius J. M. Matzke, Marjori Matzke* Institute of Plant and Microbial Biology, Academia Sinica, Taipei, Taiwan Abstract Coconut, a member of the palm family (Arecaceae), is one of the most economically important trees used by mankind. Despite its diverse morphology, coconut is recognized taxonomically as only a single species (Cocos nucifera L.). There are two major coconut varieties, tall and dwarf, the latter of which displays traits resulting from selection by humans. We report here the complete chloroplast (cp) genome of a dwarf coconut plant, and describe the gene content and organization, inverted repeat fluctuations, repeated sequence structure, and occurrence of RNA editing. Phylogenetic relationships of monocots were inferred based on 47 chloroplast protein-coding genes. Potential nodes for events of gene duplication and pseudogenization related to inverted repeat fluctuation were mapped onto the tree using parsimony criteria. We compare our findings with those from other palm species for which complete cp genome sequences are available. Citation: Huang Y-Y, Matzke AJM, Matzke M (2013) Complete Sequence and Comparative Analysis of the Chloroplast Genome of Coconut Palm (Cocos nucifera). PLoS ONE 8(8): e74736. doi:10.1371/journal.pone.0074736 Editor: Hector Candela, Universidad Miguel Herna´ndez de Elche, Spain Received June 25, 2013; Accepted August 6, 2013; Published August 30, 2013 Copyright: ß 2013 Huang et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. -

A Floristic Survey of Fair Isle
Edinburgh Research Explorer A floristic survey of Fair Isle Citation for published version: Quinteros Peñafiel, CV, Riddiford, N & Twyford, A 2017, 'A floristic survey of Fair Isle', New Journal of Botany, vol. 7, no. 2-3, pp. 101-111. https://doi.org/10.1080/20423489.2017.1393191 Digital Object Identifier (DOI): 10.1080/20423489.2017.1393191 Link: Link to publication record in Edinburgh Research Explorer Document Version: Publisher's PDF, also known as Version of record Published In: New Journal of Botany General rights Copyright for the publications made accessible via the Edinburgh Research Explorer is retained by the author(s) and / or other copyright owners and it is a condition of accessing these publications that users recognise and abide by the legal requirements associated with these rights. Take down policy The University of Edinburgh has made every reasonable effort to ensure that Edinburgh Research Explorer content complies with UK legislation. If you believe that the public display of this file breaches copyright please contact [email protected] providing details, and we will remove access to the work immediately and investigate your claim. Download date: 09. Oct. 2021 New Journal of Botany Journal of the Botanical Society of Britain & Ireland ISSN: 2042-3489 (Print) 2042-3497 (Online) Journal homepage: http://www.tandfonline.com/loi/ynjb20 A floristic survey of Fair Isle C. V. Quinteros Peñafiel, N. J. Riddiford & A. D. Twyford To cite this article: C. V. Quinteros Peñafiel, N. J. Riddiford & A. D. Twyford (2017) A floristic survey of Fair Isle, New Journal of Botany, 7:2-3, 101-111, DOI: 10.1080/20423489.2017.1393191 To link to this article: https://doi.org/10.1080/20423489.2017.1393191 © 2017 The Author(s). -

Quarterly Changes
Plant Names Database: Quarterly changes 30 November 2015 © Landcare Research New Zealand Limited 2015 This copyright work is licensed under the Creative Commons Attribution 3.0 New Zealand license. Attribution if redistributing to the public without adaptation: "Source: Landcare Research" Attribution if making an adaptation or derivative work: "Sourced from Landcare Research" http://dx.doi.org/doi:10.7931/P1Z598 CATALOGUING IN PUBLICATION Plant names database: quarterly changes [electronic resource]. – [Lincoln, Canterbury, New Zealand] : Landcare Research Manaaki Whenua, 2014- . Online resource Quarterly November 2014- ISSN 2382-2341 I.Manaaki Whenua-Landcare Research New Zealand Ltd. II. Allan Herbarium. Citation and Authorship Wilton, A.D.; Schönberger, I.; Gibb, E.S.; Boardman, K.F.; Breitwieser, I.; Cochrane, M.; Dawson, M.I.; de Pauw, B.; Fife, A.J.; Ford, K.A.; Glenny, D.S.; Heenan, P.B.; Korver, M.A.; Novis, P.M.; Redmond, D.N.; Smissen, R.D. Tawiri, K. (2015) Plant Names Database: Quarterly changes. November 2015. Lincoln, Manaaki Whenua Press. This report is generated using an automated system and is therefore authored by the staff at the Allan Herbarium who currently contribute directly to the development and maintenance of the Plant Names Database. Authors are listed alphabetically after the third author. Authors have contributed as follows: Leadership: Wilton, Heenan, Breitwieser Database editors: Wilton, Schönberger, Gibb Taxonomic and nomenclature research and review: Schönberger, Gibb, Wilton, Breitwieser, Dawson, Ford, Fife, Glenny, Heenan, Novis, Redmond, Smissen Information System development: Wilton, De Pauw, Cochrane Technical support: Boardman, Korver, Redmond, Tawiri Disclaimer The Plant Names Database is being updated every working day. We welcome suggestions for improvements, concerns, or any data errors you may find. -

A Floristic Survey of Fair Isle
Edinburgh Research Explorer A floristic survey of Fair Isle Citation for published version: Quinteros Peñafiel, CV, Riddiford, N & Twyford, A 2017, 'A floristic survey of Fair Isle', New Journal of Botany, vol. 7, no. 2-3, pp. 101-111. https://doi.org/10.1080/20423489.2017.1393191 Digital Object Identifier (DOI): 10.1080/20423489.2017.1393191 Link: Link to publication record in Edinburgh Research Explorer Document Version: Peer reviewed version Published In: New Journal of Botany General rights Copyright for the publications made accessible via the Edinburgh Research Explorer is retained by the author(s) and / or other copyright owners and it is a condition of accessing these publications that users recognise and abide by the legal requirements associated with these rights. Take down policy The University of Edinburgh has made every reasonable effort to ensure that Edinburgh Research Explorer content complies with UK legislation. If you believe that the public display of this file breaches copyright please contact [email protected] providing details, and we will remove access to the work immediately and investigate your claim. Download date: 29. Sep. 2021 1 A floristic survey of Fair Isle 2 Camila V. Quinteros Peñafiel 3 Royal Botanic Garden Edinburgh, 20A Inverleith Row, Edinburgh, EH3 5LR 4 [email protected] 5 Nick J. Riddiford 6 Schoolton, Fair Isle, Shetland, ZE2 9JU 7 [email protected] 8 Alex D. Twyford 9 Institute of Evolutionary Biology, The University of Edinburgh, Ashworth Laboratories, 10 Charlotte Auerbach Road, Edinburgh, EH9 3FL 11 [email protected] 12 A floristic survey of Fair Isle 13 14 Fair Isle is a small isolated island located off the northern tip of Great Britain. -

Directory of Azov-Black Sea Coastal Wetlands
Directory of Azov-Black Sea Coastal Wetlands Kyiv–2003 Directory of Azov-Black Sea Coastal Wetlands: Revised and updated. — Kyiv: Wetlands International, 2003. — 235 pp., 81 maps. — ISBN 90 5882 9618 Published by the Black Sea Program of Wetlands International PO Box 82, Kiev-32, 01032, Ukraine E-mail: [email protected] Editor: Gennadiy Marushevsky Editing of English text: Rosie Ounsted Lay-out: Victor Melnychuk Photos on cover: Valeriy Siokhin, Vasiliy Kostyushin The presentation of material in this report and the geographical designations employed do not imply the expres- sion of any opinion whatsoever on the part of Wetlands International concerning the legal status of any coun- try, area or territory, or concerning the delimitation of its boundaries or frontiers. The publication is supported by Wetlands International through a grant from the Ministry of Agriculture, Nature Management and Fisheries of the Netherlands and the Ministry of Foreign Affairs of the Netherlands (MATRA Fund/Programme International Nature Management) ISBN 90 5882 9618 Copyright © 2003 Wetlands International, Kyiv, Ukraine All rights reserved CONTENTS CONTENTS3 6 7 13 14 15 16 22 22 24 26 28 30 32 35 37 40 43 45 46 54 54 56 58 58 59 61 62 64 64 66 67 68 70 71 76 80 80 82 84 85 86 86 86 89 90 90 91 91 93 Contents 3 94 99 99 100 101 103 104 106 107 109 111 113 114 119 119 126 130 132 135 139 142 148 149 152 153 155 157 157 158 160 162 164 164 165 170 170 172 173 175 177 179 180 182 184 186 188 191 193 196 198 199 201 202 4 Directory of Azov-Black Sea Coastal Wetlands 203 204 207 208 209 210 212 214 214 216 218 219 220 221 222 223 224 225 226 227 230 232 233 Contents 5 EDITORIAL AND ACKNOWLEDGEMENTS This Directory is based on the national reports prepared for the Wetlands International project ‘The Importance of Black Sea Coastal Wetlands in Particular for Migratory Waterbirds’, sponsored by the Netherlands Ministry of Agriculture, Nature Management and Fisheries. -

Vascular Plants of Santa Cruz County, California
ANNOTATED CHECKLIST of the VASCULAR PLANTS of SANTA CRUZ COUNTY, CALIFORNIA SECOND EDITION Dylan Neubauer Artwork by Tim Hyland & Maps by Ben Pease CALIFORNIA NATIVE PLANT SOCIETY, SANTA CRUZ COUNTY CHAPTER Copyright © 2013 by Dylan Neubauer All rights reserved. No part of this publication may be reproduced without written permission from the author. Design & Production by Dylan Neubauer Artwork by Tim Hyland Maps by Ben Pease, Pease Press Cartography (peasepress.com) Cover photos (Eschscholzia californica & Big Willow Gulch, Swanton) by Dylan Neubauer California Native Plant Society Santa Cruz County Chapter P.O. Box 1622 Santa Cruz, CA 95061 To order, please go to www.cruzcps.org For other correspondence, write to Dylan Neubauer [email protected] ISBN: 978-0-615-85493-9 Printed on recycled paper by Community Printers, Santa Cruz, CA For Tim Forsell, who appreciates the tiny ones ... Nobody sees a flower, really— it is so small— we haven’t time, and to see takes time, like to have a friend takes time. —GEORGIA O’KEEFFE CONTENTS ~ u Acknowledgments / 1 u Santa Cruz County Map / 2–3 u Introduction / 4 u Checklist Conventions / 8 u Floristic Regions Map / 12 u Checklist Format, Checklist Symbols, & Region Codes / 13 u Checklist Lycophytes / 14 Ferns / 14 Gymnosperms / 15 Nymphaeales / 16 Magnoliids / 16 Ceratophyllales / 16 Eudicots / 16 Monocots / 61 u Appendices 1. Listed Taxa / 76 2. Endemic Taxa / 78 3. Taxa Extirpated in County / 79 4. Taxa Not Currently Recognized / 80 5. Undescribed Taxa / 82 6. Most Invasive Non-native Taxa / 83 7. Rejected Taxa / 84 8. Notes / 86 u References / 152 u Index to Families & Genera / 154 u Floristic Regions Map with USGS Quad Overlay / 166 “True science teaches, above all, to doubt and be ignorant.” —MIGUEL DE UNAMUNO 1 ~ACKNOWLEDGMENTS ~ ANY THANKS TO THE GENEROUS DONORS without whom this publication would not M have been possible—and to the numerous individuals, organizations, insti- tutions, and agencies that so willingly gave of their time and expertise. -

Dissertation Pdf .Odt
The floodplain meadows of Soomaa National Park, Estonia Vegetation – Dispersal – Regeneration Dissertation zur Erlangung des Doktorgrades (Dr. rer. nat.) der Naturwissenschaftlichen Fakultät III – Biologie und vorklinische Medizin – der Universität Regensburg vorgelegt von Jaan Palisaar aus Kiel Edertal, April 2006 Promotionsgesuch eingereicht am 10. April 2006 Tag der mündlichen Prüfung 26. Juli 2006 Die Arbeit wurde angeleitet von Prof. Dr. Peter Poschlod Prüfungsausschuß: Prof. Dr. Jürgen Heinze Prof. Dr. Peter Poschlod Prof. Dr. Karl-Georg Bernhardt Prof. Dr. Christoph Oberprieler Contents List of figures.........................................................................................................................III List of tables...........................................................................................................................VI Acknowledgments................................................................................................................IX A. Foreword.............................................................................................................................1 B. Study area............................................................................................................................3 1 Physical setting...............................................................................................................3 2 Land use...........................................................................................................................8 C. Vegetation -
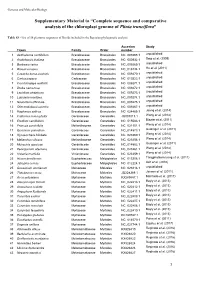
Complete Sequence and Comparative Analysis of the Chloroplast Genome of Plinia Trunciflora”
Genetics and Molecular Biology Supplementary Material to “Complete sequence and comparative analysis of the chloroplast genome of Plinia trunciflora” Table S3 - List of 56 plastome sequences of Rosids included in the Bayesian phylogenetic analysis. Accesion Study Taxon Family Order number 1 Aethionema cordifolium Brassicaceae Brassicales NC_009265.1 unpublished 2 Arabidopsis thaliana Brassicaceae Brassicales NC_000932.1 Sato et al. (1999) 3 Barbarea verna Brassicaceae Brassicales NC_009269.1 unpublished 4 Brassica napus Brassicaceae Brassicales NC_016734.1 Hu et al. (2011) 5 Capsella bursa-pastoris Brassicaceae Brassicales NC_009270.1 unpublished 6 Carica papaya Caricaceae Brassicales NC_010323.1 unpublished 7 Crucihimalaya wallichii Brassicaceae Brassicales NC_009271.1 unpublished 8 Draba nemorosa Brassicaceae Brassicales NC_009272.1 unpublished 9 Lepidium virginicum Brassicaceae Brassicales NC_009273.1 unpublished 10 Lobularia maritima Brassicaceae Brassicales NC_009274.1 unpublished 11 Nasturtium officinale Brassicaceae Brassicales NC_009275.1 unpublished 12 Olimarabidopsis pumila Brassicaceae Brassicales NC_009267.1 unpublished 13 Raphanus sativus Brassicaceae Brassicales NC_024469.1 Jeong et al. (2014) 14 California macrophylla Geraniaceae Geraniales JQ031013.1 Weng et al. (2014) 15 Erodium carvifolium Geraniaceae Geraniales NC_015083.1 Blazier et al. (2011) 16 Francoa sonchifolia Melianthaceae Geraniales NC_021101.1 Weng et al. (2014) 17 Geranium palmatum Geraniaceae Geraniales NC_014573.1 Guisinger et al. (2011) 18 Hypseocharis bilobate Geraniaceae Geraniales NC_023260.1 Weng et al. (2014) 19 Melianthus villosus Melianthaceae Geraniales NC_023256.1 Weng et al. (2014) 20 Monsonia speciose Geraniaceae Geraniales NC_014582.1 Guisinger et al. (2011) 21 Pelargonium alternans Geraniaceae Geraniales NC_023261.1 Weng et al. (2014) 22 Viviania marifolia Vivianiaceae Geraniales NC_023259.1 Weng et al. (2014) 23 Hevea brasiliensis Euphorbiaceae Malpighiales NC_015308.1 Tangphatsornruang et al. -

Downloaded from Genbank on That Full Plastid Genomes Are Not Sufficient to Reject Al- February 28, 2012
Ruhfel et al. BMC Evolutionary Biology 2014, 14:23 http://www.biomedcentral.com/1471-2148/14/23 RESEARCH ARTICLE Open Access From algae to angiosperms–inferring the phylogeny of green plants (Viridiplantae) from 360 plastid genomes Brad R Ruhfel1*, Matthew A Gitzendanner2,3,4, Pamela S Soltis3,4, Douglas E Soltis2,3,4 and J Gordon Burleigh2,4 Abstract Background: Next-generation sequencing has provided a wealth of plastid genome sequence data from an increasingly diverse set of green plants (Viridiplantae). Although these data have helped resolve the phylogeny of numerous clades (e.g., green algae, angiosperms, and gymnosperms), their utility for inferring relationships across all green plants is uncertain. Viridiplantae originated 700-1500 million years ago and may comprise as many as 500,000 species. This clade represents a major source of photosynthetic carbon and contains an immense diversity of life forms, including some of the smallest and largest eukaryotes. Here we explore the limits and challenges of inferring a comprehensive green plant phylogeny from available complete or nearly complete plastid genome sequence data. Results: We assembled protein-coding sequence data for 78 genes from 360 diverse green plant taxa with complete or nearly complete plastid genome sequences available from GenBank. Phylogenetic analyses of the plastid data recovered well-supported backbone relationships and strong support for relationships that were not observed in previous analyses of major subclades within Viridiplantae. However, there also is evidence of systematic error in some analyses. In several instances we obtained strongly supported but conflicting topologies from analyses of nucleotides versus amino acid characters, and the considerable variation in GC content among lineages and within single genomes affected the phylogenetic placement of several taxa. -

In Situ Analysis of Grass Cell Wall Polysaccharides
In situ analysis of grass cell wall polysaccharides By Jie Xue Submitted in accordance with the requirements for the degree of Doctor of Philosophy The University of Leeds Faculty of Biological Sciences November 2013 This copy has been supplied on the understanding that it is copyright material and that no quotation from the thesis may be published without proper acknowledgement. © 2013 The University of Leeds and Jie Xue The candidate confirms that all the work submitted in this thesis is her own. The candidate confirms that appropriate credit has been given within the thesis where reference has been made to the work of others. Some of the work presented in Chapters 3 and 5 appear in the following publication: Jie Xue, Maurice Bosch, and J. Paul Knox, 2013. Heterogeneity and glycan masking of cell wall microstructures in the stems of Miscanthus x giganteus, and its parents M. sinensis and M. sacchariflorus. Plos ONE 8, e82114. “Happy is the man who is living by his hobby” George Bernard Shaw, 1856-1950 Acknowledgements Firstly I am very grateful to my supervisor, Professor Paul Knox, gave me a chance here for my PhD study, and also for all his help, instructions, guidance and patience for my study. The knowledgeable man (responsible for my interest in cell walls) taught me how to do an excellent cell wall research, how to produce a good scientific work and how to be a responsible scientist. Susan Marcus, the most kind woman in the Knox lab, for being taking very good care of me during these past three years, and also for her excellent lab techniques.