Genetic Diversity in Leavenworthia Populations with Different Inbreeding Levels
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Federal Register/Vol. 84, No. 119/Thursday, June 20, 2019/Notices
28850 Federal Register / Vol. 84, No. 119 / Thursday, June 20, 2019 / Notices or speech-impaired individuals may status reviews of 53 species under the Relay Service at 800–877–8339 for TTY access this number through TTY by Endangered Species Act, as amended. A assistance. calling the toll-free Federal Relay 5-year review is an assessment of the SUPPLEMENTARY INFORMATION: Service at 800–877–8339. best scientific and commercial data Dated: June 14, 2019. available at the time of the review. We Why do we conduct 5-year reviews? are requesting submission of Brian D. Montgomery, Under the Endangered Species Act of Acting Deputy Secretary. information that has become available since the last reviews of these species. 1973, as amended (ESA; 16 U.S.C. 1531 [FR Doc. 2019–13146 Filed 6–19–19; 8:45 am] et seq.), we maintain lists of endangered BILLING CODE 4210–67–P DATES: To allow us adequate time to and threatened wildlife and plant conduct these reviews, we must receive species in title 50 of the Code of Federal your comments or information on or Regulations (CFR) at 50 CFR 17.11 (for DEPARTMENT OF THE INTERIOR before August 19, 2019. However, we wildlife) and 17.12 (for plants: List). will continue to accept new information Section 4(c)(2)(A) of the ESA requires us Fish and Wildlife Service about any listed species at any time. to review each listed species’ status at least once every 5 years. Our regulations [FWS–R4–ES–2019–N037; ADDRESSES: For instructions on how to FXES11130900000C2–190–FF09E32000] submit information and review at 50 CFR 424.21 require that we publish a notice in the Federal Register Endangered and Threatened Wildlife information that we receive on these species, see Request for New announcing those species under active and Plants; Initiation of 5-Year Status review. -

Environmental Report (ER) (TVA 2003) in Conjunction with Its Application for Renewal of the BFN Ols, As Provided for by the Following NRC Regulations
Biological Assessment Browns Ferry Nuclear Power Plant License Renewal Review Limestone County, Alabama October 2004 Docket Numbers 50-259, 50-260, and 50-296 U.S. Nuclear Regulatory Commission Rockville, Maryland Biological Assessment of the Potential Effects on Endangered or Threatened Species from the Proposed License Renewal for the Browns Ferry Nuclear Plant 1.0 Introduction The U.S. Nuclear Regulatory Commission (NRC) licenses the operation of domestic nuclear power plants in accordance with the Atomic Energy Act of 1954, as amended, and NRC implementing regulations. The Tennessee Valley Authority (TVA) operates Browns Ferry Nuclear Power Plant, Units 1, 2, and 3 (BFN) pursuant to NRC operating license (OL) numbers DPR-33, DPR-52, DPR-68, which expire on December 20, 2013, June 28, 2014, and July 2, 2016, respectively. TVA has prepared an Environmental Report (ER) (TVA 2003) in conjunction with its application for renewal of the BFN OLs, as provided for by the following NRC regulations: C Title 10 of the Code of Federal Regulations, Part 54, “Requirements for Renewal of Operating Licenses for Nuclear Power Plants,” Section 54.23, Contents of application - environmental information (10 CFR 54.23). C Title 10 of the Code of Federal Regulations, Part 51, “Environmental Protection Regulations for Domestic Licensing and Related Regulatory Functions,” Section 51.53, Postconstruction environmental reports, Subsection 51.53(c), Operating license renewal stage (10 CFR 51.53(c)). The renewed OLs would allow up to 20 additional years of plant operation beyond the current licensed operating term. No major refurbishment or replacement of important systems, structures, or components are expected during the 20-year BFN license renewal term. -

Heterochrony of Floral and Mating System Characters Between Nicotiana Longiflora and N
HETEROCHRONY OF FLORAL AND MATING SYSTEM CHARACTERS BETWEEN NICOTIANA LONGIFLORA AND N. PLUMBAGINIFOLIA A Thesis presented to the Faculty of the Graduate School at the University of Missouri-Columbia In Partial Fulfillment of the Requirements for the Degree Master of Arts by JACOB W. SOULE Dr. Timothy P. Holtsford, Thesis Supervisor MAY 2007 The undersigned, appointed by the dean of the Graduate School, have examined the thesis entitled HETEROCHRONY OF FLORAL AND MATING SYSTEM CHARACTERS BETWEEN NICOTIANA LONGIFLORA AND N. PLUMBAGINIFOLIA presented by Jacob W. Soule, a candidate for the degree of Master of Arts, and hereby certify that, in their opinion, it is worthy of acceptance. Professor Timothy P. Holtsford Professor J. Chris Pires Professor Bruce A. McClure ACKNOWLEDGEMENTS This thesis would not have been possible without my advisor Dr. Tim Holtsford. His insight and thoughtful comments helped me to bring this to completion. He helped me focus my project into something worthwhile. I had a lot of help from my lab mates Rainee Kaczorowski, Sherry Ellberg, Dulce Figueroa-Castro and Chris Lee; their discussions helped me to get past many difficulties for which I am grateful, as well as for their willing help with watering and care of plants. I would like to thank Paul Walker for his enthusiastic help with lab work and for the hours he spent measuring flowers for plastochron calibrations. The terrific feedback given to me by my committee members Dr. J. Chris Pires and Dr. Bruce A. McClure greatly improved the final product. Finally, I never would have made it through this without the love and support of my parents, Robert and Judith Soule, and of my fiancée Danielle A. -

How Does Genome Size Affect the Evolution of Pollen Tube Growth Rate, a Haploid Performance Trait?
Manuscript bioRxiv preprint doi: https://doi.org/10.1101/462663; this version postedClick April here18, 2019. to The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv aaccess/download;Manuscript;PTGR.genome.evolution.15April20 license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Effects of genome size on pollen performance 2 3 4 5 How does genome size affect the evolution of pollen tube growth rate, a haploid 6 performance trait? 7 8 9 10 11 John B. Reese1,2 and Joseph H. Williams2 12 Department of Ecology and Evolutionary Biology, University of Tennessee, Knoxville, TN 13 37996, U.S.A. 14 15 16 17 1Author for correspondence: 18 John B. Reese 19 Tel: 865 974 9371 20 Email: [email protected] 21 1 bioRxiv preprint doi: https://doi.org/10.1101/462663; this version posted April 18, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 22 ABSTRACT 23 Premise of the Study – Male gametophytes of most seed plants deliver sperm to eggs via a 24 pollen tube. Pollen tube growth rates (PTGRs) of angiosperms are exceptionally rapid, a pattern 25 attributed to more effective haploid selection under stronger pollen competition. Paradoxically, 26 whole genome duplication (WGD) has been common in angiosperms but rare in gymnosperms. -

Evolutionary Genetics of Core Eudicot Inflorescence and Flower Development
® International Journal of Plant Developmental Biology ©2010 Global Science Books Evolutionary Genetics of Core Eudicot Inflorescence and Flower Development Jill C. Preston* Department of Ecology and Evolutionary Biology, University of Kansas, 1200 Sunnyside Avenue, Lawrence, KS 66045, USA Corresponding author : * [email protected] ABSTRACT The genetic basis of flowering is best understood in the model core eudicot species Arabidopsis thaliana (Brassicaceae), and involves the genetic reprogramming of shoot apical meristems, ending in the production of flowers. Although inflorescences and flowers of core eudicots share a common ground plan, variation in architecture, shape and ornamentation suggests repeated modifications to this ancestral plan. Comparative studies, primarily in Brassicaceae and Leguminoseae (rosids), and Asteraceae, Plantaginaceae and Solanaceae (asterids), have revealed a common developmental framework for flowering across core eudicots. This serves as a basis for understanding genetic changes that underlie the diversification of inflorescence and floral form. Recent work is starting to reveal the relative importance of regulatory versus protein coding changes in genes involved in diversification of inflorescence and flower development across core eudicots. Furthermore, these studies highlight the importance of phylogenetic history for understanding functional conservation of duplicated genes. _____________________________________________________________________________________________________________ Keywords: Antirrhinum -

Biological Evaluation of Proposed, Threatened, Endangered and Sensitive Species
Biological Evaluation of Proposed, Threatened, Endangered and Sensitive Species Wildlife Habitat Improvement and Fuels Reduction Project Proposed Action within Winston County, Alabama Responsible Agency: USDA Forest Service National Forests in Alabama William B. Bankhead Ranger District Contact: Deciding Officer: District Ranger Glen D. Gaines Biological Evaluation Preparer: Biological Scientist Allison Cochran PO Box 278 Double Springs, Alabama 35553 Telephone 205-489-5111 FAX 205-489-3427 E-mail [email protected] [email protected] Type of Document Categorical Exclusion – BE Summary The proposed project will reduce midstory and understory trees and shrubs in two sites, totaling approximately 47 acres, noted on the attached maps. The project sites are located in the Black Pond and Hickory Grove communities. They are found in Forest Service management compartments 163 and 19. The sites proposed for treatment are loblolly pine stands. They were thinned in 2005 and 2006, respectively. Selected vegetation between 1 inch and 6 inches DBH will be removed in upland pine- dominant habitat. In compartment 163, the result will be an open pine stand with reduced fuel loading and advanced hardwood regeneration. In compartment 19, the result will be an open pine stand with reduced fuel loading. The result will allow for restoration and maintenance of native forest communities, including upland oak-hickory forest in compartment 163 and fire dependent pine woodlands in compartment 19. The purpose and need for the project is to improve wildlife habitat, improve conditions for native upland plants, restore and maintain native forest communities, and to decrease the risk of catastrophic wildfires by reducing fuels. -

Fleshy-Fruit Ladecress
Fleshy-fruit Gladecress GP07072016 IDENTIFICATION: Fleshy-fruit gladecress, Leavenworthia crassa, is a member of the mustard family (Brassicaceae) that inhabits cedar glades at seven known sites in Alabama. It was listed as an endangered species on September 2, 2014. A winter annual with leaves that are mostly basal, forming a rosette, and entire to very deeply pinnately lobed or divided and up to about three inches in length. The plant usually grows from four to 12 inches tall. Flowers are typically about ½ inch in length and either yellow with orange or white with orange in color. Typically blooms in mid-March through mid- April. Both color variations may be seen in a single population. Dark brown, winged seeds are within a globe-shaped or slightly oblong fruit approximately ½ inch in diameter. HABITAT: This species is associated with areas often referred to as “glades” or “cedar glades.” Much of the cedar glade habitat in north Alabama is in a degraded condition, and populations of fleshy-fruit gladecress, in many cases, persist in glade-like remnants exhibiting various degrees of disturbance including pastures, roadside rights-of-way, and cultivated or plowed fields. The limestone outcrops, gravel, and shallow soils present in cedar glades and glade-like remnants provide space for individual and population growth of fleshy-fruit gladecress by regulating the encroachment of herbaceous and woody vegetation that would exclude fleshy-fruit gladecress from plant communities found on deeper soils. Eastern red cedar is often present on the glade edges in deeper soils. Wide variations in soil moisture content are not uncommon in typical “glade” habitats. -
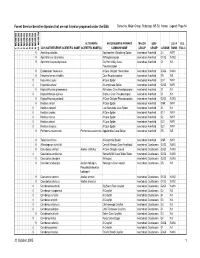
Sensitive Species That Are Not Listed Or Proposed Under the ESA Sorted By: Major Group, Subgroup, NS Sci
Forest Service Sensitive Species that are not listed or proposed under the ESA Sorted by: Major Group, Subgroup, NS Sci. Name; Legend: Page 94 REGION 10 REGION 1 REGION 2 REGION 3 REGION 4 REGION 5 REGION 6 REGION 8 REGION 9 ALTERNATE NATURESERVE PRIMARY MAJOR SUB- U.S. N U.S. 2005 NATURESERVE SCIENTIFIC NAME SCIENTIFIC NAME(S) COMMON NAME GROUP GROUP G RANK RANK ESA C 9 Anahita punctulata Southeastern Wandering Spider Invertebrate Arachnid G4 NNR 9 Apochthonius indianensis A Pseudoscorpion Invertebrate Arachnid G1G2 N1N2 9 Apochthonius paucispinosus Dry Fork Valley Cave Invertebrate Arachnid G1 N1 Pseudoscorpion 9 Erebomaster flavescens A Cave Obligate Harvestman Invertebrate Arachnid G3G4 N3N4 9 Hesperochernes mirabilis Cave Psuedoscorpion Invertebrate Arachnid G5 N5 8 Hypochilus coylei A Cave Spider Invertebrate Arachnid G3? NNR 8 Hypochilus sheari A Lampshade Spider Invertebrate Arachnid G2G3 NNR 9 Kleptochthonius griseomanus An Indiana Cave Pseudoscorpion Invertebrate Arachnid G1 N1 8 Kleptochthonius orpheus Orpheus Cave Pseudoscorpion Invertebrate Arachnid G1 N1 9 Kleptochthonius packardi A Cave Obligate Pseudoscorpion Invertebrate Arachnid G2G3 N2N3 9 Nesticus carteri A Cave Spider Invertebrate Arachnid GNR NNR 8 Nesticus cooperi Lost Nantahala Cave Spider Invertebrate Arachnid G1 N1 8 Nesticus crosbyi A Cave Spider Invertebrate Arachnid G1? NNR 8 Nesticus mimus A Cave Spider Invertebrate Arachnid G2 NNR 8 Nesticus sheari A Cave Spider Invertebrate Arachnid G2? NNR 8 Nesticus silvanus A Cave Spider Invertebrate Arachnid G2? NNR -

USFWS Consultation on Transmission System Right-Of-Way Program
United States Department of the Interior FISH AND WILDLIFE SERVICE Tennessee ES Office 446 Neal Street Cookeville, Tennessee 38501 December 18, 2018 Mr. John T. Baxter Manager, Biological Compliance Tennessee Valley Authority 400 West Summit Hill Drive Knoxville, TN 37902 Re: FWS #2018-F-0958; Programmatic Consultation for Right-of-Way Vegetation Management that May Affect Endangered or Threatened Plants in the Tennessee Valley Authority Service Area Dear Mr. Baxter: This letter acknowledges the U.S. Fish and Wildlife Service’s (Service) November 21, 2018, receipt of your November 19, 2015, letter requesting initiation of formal section 7 consultation under the Endangered Species Act (Act). The consultation concerns the possible effects of your proposed Programmatic Strategy for Right-of-Way Vegetation Management that May Affect Endangered or Threatened Plants in the Tennessee Valley Authority Service Area (TVA) (the Proposed Action) on 18 federally listed plants, including: • Price's potato-bean (Apios priceana) • Braun's rock-cress (Arabis perstellata) • Pyne's ground plum (Astragalus bibullatus) • Morefield's leather-flower (Clematis morefieldii) • Alabama leather flower (Clematis socialis) • leafy prairie-clover (Dalea foliosa) • whorled sunflower (Helianthus verticillatus) • small whorled pogonia (Isotria medeoloides) • fleshy-fruit gladecress (Leavenworthia crassa) • lyre-leaf bladderpod (Lesquerella lyrata) • Spring Creek bladderpod (Lesquerella perforata) • Mohr's Barbara's buttons (Marshallia mohrii) • Cumberland sandwort (Minuartia cumberlandensis) • Short’s bladderpod (Physaria globosa) • white fringeless orchid (Platanthera integrilabia) • green pitcher plant (Sarracenia oreophila) • large-flowered skullcap (Scutellaria montana) • Tennessee yellow-eyed grass (Xyris tennesseensis) Listed species (LE=listed as endangered; LT=listed as threatened) and designated critical habitats (DCH) that TVA has determined the proposed Action is not likely to adversely affect (NLAA). -

Helianthus Verticillatus (Whorled Sunflower), and Leavenworthia Crassa (Fleshy-Fruit Gladecress); Final Rule
Vol. 79 Tuesday, No. 165 August 26, 2014 Part II Department of the Interior Fish and Wildlife Service 50 CFR Part 17 Endangered and Threatened Wildlife and Plants; Designation of Critical Habitat for Physaria globosa (Short’s bladderpod), Helianthus verticillatus (whorled sunflower), and Leavenworthia crassa (fleshy-fruit gladecress); Final Rule VerDate Mar<15>2010 17:29 Aug 25, 2014 Jkt 232001 PO 00000 Frm 00001 Fmt 4717 Sfmt 4717 E:\FR\FM\26AUR2.SGM 26AUR2 tkelley on DSK3SPTVN1PROD with RULES2 50990 Federal Register / Vol. 79, No. 165 / Tuesday, August 26, 2014 / Rules and Regulations DEPARTMENT OF THE INTERIOR available at the Fish and Wildlife opinions from five knowledgeable Service Web site and Field Office set out individuals with scientific expertise to Fish and Wildlife Service above, and may also be included in the review our technical assumptions, preamble and at http:// analysis, and whether or not we had 50 CFR Part 17 www.regulations.gov. used the best available information. [Docket No. FWS–R4–ES–2013–0086; FOR FURTHER INFORMATION CONTACT: These peer reviewers generally 4500030114] Mary E. Jennings, Field Supervisor, U.S. concurred with our methods and Fish and Wildlife Service, Tennessee conclusions and provided additional RIN 1018–AZ60 information, clarifications, and Ecological Services Fish and Wildlife suggestions to improve this final rule. Endangered and Threatened Wildlife Office, (see ADDRESSES above). Persons Information we received from peer who use a telecommunications device and Plants; Designation of Critical review is incorporated in this final for the deaf (TDD) may call the Federal Habitat for Physaria globosa (Short’s revised designation. -

The Vascular Flora of the Natchez Trace Parkway
THE VASCULAR FLORA OF THE NATCHEZ TRACE PARKWAY (Franklin, Tennessee to Natchez, Mississippi) Results of a Floristic Inventory August 2004 - August 2006 © Dale A. Kruse, 2007 © Dale A. Kruse 2007 DATE SUBMITTED 28 February 2008 PRINCIPLE INVESTIGATORS Stephan L. Hatch Dale A. Kruse S. M. Tracy Herbarium (TAES), Texas A & M University 2138 TAMU, College Station, Texas 77843-2138 SUBMITTED TO Gulf Coast Inventory and Monitoring Network Lafayette, Louisiana CONTRACT NUMBER J2115040013 EXECUTIVE SUMMARY The “Natchez Trace” has played an important role in transportation, trade, and communication in the region since pre-historic times. As the development and use of steamboats along the Mississippi River increased, travel on the Trace diminished and the route began to be reclaimed by nature. A renewed interest in the Trace began during, and following, the Great Depression. In the early 1930’s, then Mississippi congressman T. J. Busby promoted interest in the Trace from a historical perspective and also as an opportunity for employment in the area. Legislation was introduced by Busby to conduct a survey of the Trace and in 1936 actual construction of the modern roadway began. Development of the present Natchez Trace Parkway (NATR) which follows portions of the original route has continued since that time. The last segment of the NATR was completed in 2005. The federal lands that comprise the modern route total about 52,000 acres in 25 counties through the states of Alabama, Mississippi, and Tennessee. The route, about 445 miles long, is a manicured parkway with numerous associated rest stops, parks, and monuments. Current land use along the NATR includes upland forest, mesic prairie, wetland prairie, forested wetlands, interspersed with numerous small agricultural croplands. -

Short's Bladderpod
30792 Federal Register / Vol. 79, No. 103 / Thursday, May 29, 2014 / Proposed Rules DEPARTMENT OF THE INTERIOR Field Office (see FOR FURTHER (2) Specific information on: INFORMATION CONTACT). (a) The distribution of Short’s Fish and Wildlife Service Written Comments: You may submit bladderpod, whorled sunflower, and written comments by one of the fleshy-fruit gladecress; 50 CFR Part 17 following methods: (b) The amount and distribution of habitat for Short’s bladderpod, whorled [Docket No. FWS–R4–ES–2013–0086; (1) Electronically: Go to the Federal 4500030113] eRulemaking Portal: http:// sunflower, and fleshy-fruit gladecress; www.regulations.gov. Submit comments and RIN 1018–AZ60 on the critical habitat proposal and (c) What areas occupied by the associated DEA by searching for FWS– species at the time of listing that contain Endangered and Threatened Wildlife R4–ES–2013–0086, which is the docket features essential for the conservation of and Plants; Designation of Critical number for this rulemaking. the species we should include in the Habitat for Physaria globosa (Short’s (2) By hard copy: Submit comments designation and why, and Bladderpod), Helianthus verticillatus on the critical habitat proposal and (d) What areas not occupied at the (Whorled Sunflower), and associated DEA by U.S. mail or hand- time of listing are essential to the Leavenworthia crassa (Fleshy-Fruit delivery to: Public Comments conservation of the species and why. Gladecress) Processing, Attn: FWS–R4–ES–2013– (3) Land use designations and current AGENCY: Fish and Wildlife Service, 0086; Division of Policy and Directives or planned activities in the subject areas Interior.