Gene Discovery and Differential Expression
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Wnt Signaling Pathway Is Involved in the Regulation of Phagocytosis Of
OPEN The Wnt signaling pathway is involved in SUBJECT AREAS: the regulation of phagocytosis of virus in PHAGOCYTES CELL BIOLOGY Drosophila CYTOSKELETON Fei Zhu1,2 & Xiaobo Zhang1 CELLULAR MICROBIOLOGY 1Key Laboratory of Conservation Biology for Endangered Wildlife of Ministry of Education, Key Laboratory of Animal Virology of 2 Received Ministry of Agriculture and College of Life Sciences, Zhejiang University, Hangzhou 310058, China, College of Animal Science 1 February 2013 and Technology, Zhejiang Agriculture and Forestry University, Hangzhou 311300, China. Accepted 3 June 2013 Phagocytosis is crucial for triggering host defenses against invading pathogens in animals. However, the receptors on phagocyte surface required for phagocytosis of virus have not been extensively explored. This Published study demonstrated that white spot syndrome virus (WSSV), a major pathogen of shrimp, could be engulfed 25 June 2013 but not digested by Drosophila S2 cells, indicating that the virus was not recognized and taken up by a pathway that was silent and would not activate the phagosome maturation and digestion pathway. The results showed that the activation of receptors on S2 cell surface by lipopolysaccharide or peptidoglycan resulted in the phagocytosis of S2 cells against WSSV virions. Gene expression profiles revealed that the Correspondence and dally-mediated Wnt signaling pathway was involved in S2 phagocytosis. Further data showed that the Wnt requests for materials signaling pathway played an essential role in phagocytosis. Therefore, our study contributed novel insight should be addressed to into the molecular mechanism of phagocytosis in animals. X.B.Z. (zxb0812@zju. edu.cn) hagocytosis, a highly conserved process, is crucial for the immune responses of animals and it allows for rapid engulfment of pathogens and apoptotic cells by specialized phagocytes1–5. -

Transcriptional Regulation by Extracellular Signals 209
Cell, Vol. 80, 199-211, January 27, 1995, Copyright © 1995 by Cell Press Transcriptional Regulation Review by Extracellular Signals: Mechanisms and Specificity Caroline S. Hill and Richard Treisman Nuclear Translocation Transcription Laboratory In principle, regulated nuclear localization of transcription Imperial Cancer Research Fund factors can involve regulated activity of either nuclear lo- Lincoln's Inn Fields calization signals (NLSs) or cytoplasmic retention signals, London WC2A 3PX although no well-characterized case of the latter has yet England been reported. N LS activity, which is generally dependent on short regions of basic amino acids, can be regulated either by masking mechanisms or by phosphorylations Changes in cell behavior induced by extracellular signal- within the NLS itself (Hunter and Karin, 1992). For exam- ing molecules such as growth factors and cytokines re- ple, association with an inhibitory subunit masks the NLS quire execution of a complex program of transcriptional of NF-KB and its relatives (Figure 1; for review see Beg events. While the route followed by the intracellular signal and Baldwin, 1993), while an intramolecular mechanism from the cell membrane to its transcription factor targets may mask NLS activity in the heat shock regulatory factor can be traced in an increasing number of cases, how the HSF2 (Sheldon and Kingston, 1993). When transcription specificity of the transcriptional response of the cell to factor localization is dependent on regulated NLS activity, different stimuli is determined is much less clear. How- linkage to a constitutively acting NLS may be sufficient to ever, it is possible to understand at least in principle how render nuclear localization independent of signaling (Beg different stimuli can activate the same signal pathway yet et al., 1992). -

NF-Κb Modifies the Mammalian Circadian Clock Through Interaction with the Core Clock Protein BMAL1
bioRxiv preprint doi: https://doi.org/10.1101/2020.09.06.285254; this version posted September 6, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. NF-κB modifies the mammalian circadian clock through interaction with the core clock protein BMAL1 Yang Shen1, Wei Wang2, Mehari Endale1, Lauren J. Francey3, Rachel L. Harold4, David W. Hammers5, Zhiguang Huo6, Carrie L. Partch4, John B. Hogenesch3, Zhao-Hui Wu2, and Andrew C. Liu1* 1Department of Physiology and Functional Genomics, University of Florida College of Medicine; 2Department of Radiation Oncology and Center for Cancer Research, University of Tennessee Health Science Center; 3Divisions of Human Genetics and Immunobiology, Cincinnati Children's Hospital Medical Center; 4Department of Chemistry and Biochemistry, University of California Santa Cruz; 5Department of Pharmacology and Therapeutics, University of Florida College of Medicine; 6Department of Biostatistics, College of Public Health & Health Professions, University of Florida College of Medicine Running title: NF-κB modifies circadian clock * Correspondence: Andrew C. Liu, [email protected] Keywords: Circadian clock, inflammation, NF-κB, suprachiasmatic nucleus (SCN), BMAL1 Abbreviations: NF-κB, nuclear factor kappa B; SCN, suprachiasmatic nucleus; IKK, IκB kinase; IκBα, inhibitor of NF-κB; Luc, luciferase; RHD, Rel homology domain; CRD, C-terminal oscillation regulatory domain, TAD, transactivation domain 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.09.06.285254; this version posted September 6, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. -

Dynamic Regulation of Innate Immune Responses in Drosophila by Senju-Mediated Glycosylation
Dynamic regulation of innate immune responses in Drosophila by Senju-mediated glycosylation Miki Yamamoto-Hinoa, Masatoshi Muraokab, Shu Kondoc, Ryu Uedac, Hideyuki Okanod, and Satoshi Gotoa,d,1 aDepartment of Life Science, Rikkyo University, Tokyo 171-8501, Japan; bStem Cell Project Group, Tokyo Metropolitan Institute of Medical Science, Tokyo 156-8506, Japan; cInvertebrate Genetics Laboratory, Genetic Strains Research Center, National Institute of Genetics, Mishima 411-8540, Japan; and dDepartment of Physiology, Keio University School of Medicine, Tokyo 160-8582, Japan Edited by Norbert Perrimon, Howard Hughes Medical Institute, Harvard Medical School, Boston, MA, and approved March 30, 2015 (received for review December 22, 2014) The innate immune system is the first line of defense encountered by Most cell surface and secreted proteins are glycosylated in the invading pathogens. Delayed and/or inadequate innate immune endoplasmic reticulum (ER) and Golgi apparatus. Glycosylation responses can result in failure to combat pathogens, whereas ex- requires glycosyltransferases and nucleotide sugar substrates cessive and/or inappropriate responses cause runaway inflamma- that are synthesized in the cytosol and nucleus. The nucleotide tion. Therefore, immune responses are tightly regulated from sugars must be transported into the ER and Golgi lumens by initiation to resolution and are repressed during the steady state. It is nucleotide sugar transporters (NSTs) (6). On arrival, they are used well known that glycans presented on pathogens play important by glycosyltransferases. The Drosophila genome encodes at least roles in pathogen recognition and the interactions between host 10 NSTs, which transport different subsets of nucleotide sugars. molecules and microbes; however, the function of glycans of host Studies of mutated NST genes show that protein glycosylation organisms in innate immune responses is less well known. -
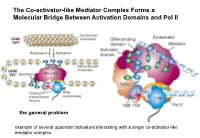
The Co-Activator-Like Mediator Complex Forms a Molecular Bridge Between Activation Domains and Pol II
The Co-activator-like Mediator Complex Forms a Molecular Bridge Between Activation Domains and Pol II Co-activator the general problem example of several upstream activators interacting with a single co-activator-like mediator complex Transcription Regulation And Gene Expression in Eukaryotes FS 2016 Graduate Course G2 P. Matthias and RG Clerc Pharmazentrum Hörsaal 2 16h15-18h00 REGULATORY MECHANISMS OF TRANSCRIPTION FACTOR FUNCTION •Protein synthesized •Protein phosphorylated •Ligand binding •Release inhibitor •Change partner, etc RG Clerc April 6. 2016 Transcription factors as final effectors of the cellular signaling cascade Regulatory Mechanisms of Transcription Factor Function Genes X. Lewin B. editor Regulatory Mechanisms of Transcription Factor Function CREB FOXO NFAT Genes X. Lewin B. editor Body plan is constructed through interactions of the developmentally regulated homeotic gene expression anterior early expression posterior late expression high RA response low RA response hindbrain trunk Alexander T and Krumlauf R. Ann.Rev.Cell.Dev.Biol: 25_431 (2009) Hoxb transcription factors mRNA distribution along the AP axis A staggered expression of the anterior border within somites is a property of the physical ordering along the chromosome: the colinearity Alexander T and Krumlauf R. Ann.Rev.Cell.Dev.Biol: 25_431 (2009) Transcription control of the Hox genes: insight into colinear activation P A p p p p a p a p a a a a Tarchini B and Duboule D. Dev.Cell 10_93 (2006) correlation between linear arrangement along the chromosome and spatial -

Amphioxus TLR Signaling Adaptors Act As Negative Regulators In
Novel Toll/IL-1 Receptor Homologous Region Adaptors Act as Negative Regulators in Amphioxus TLR Signaling This information is current as Jian Peng, Xin Tao, Rui Li, Jingru Hu, Jie Ruan, Ruihua of September 29, 2021. Wang, Manyi Yang, Rirong Yang, Xiangru Dong, Shangwu Chen, Anlong Xu and Shaochun Yuan J Immunol published online 31 August 2015 http://www.jimmunol.org/content/early/2015/08/30/jimmun ol.1403003 Downloaded from Supplementary http://www.jimmunol.org/content/suppl/2015/08/30/jimmunol.140300 Material 3.DCSupplemental http://www.jimmunol.org/ Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication by guest on September 29, 2021 *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2015 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. Published August 31, 2015, doi:10.4049/jimmunol.1403003 The Journal of Immunology Novel Toll/IL-1 Receptor Homologous Region Adaptors Act as Negative Regulators in Amphioxus TLR Signaling Jian Peng,*,1 Xin Tao,* Rui Li,* Jingru Hu,* Jie Ruan,* Ruihua Wang,* Manyi Yang,* Rirong Yang,* Xiangru Dong,* Shangwu Chen,* Anlong Xu,*,† and Shaochun Yuan* Studies have shown that the basal chordate amphioxus possesses an extraordinarily complex TLR system, including 39 TLRs and at least 40 Toll/IL-1R homologous region (TIR) adaptors. -

The C-Rel Transcription Factor and B-Cell Proliferation: a Deal with the Devil
Oncogene (2004) 23, 2275–2286 & 2004 Nature Publishing Group All rights reserved 0950-9232/04 $25.00 www.nature.com/onc REVIEW The c-Rel transcription factor and B-cell proliferation: a deal with the devil Thomas D Gilmore*,1, Demetrios Kalaitzidis1, Mei-Chih Liang1 and Daniel T Starczynowski1 1Department of Biology, Boston University, 5 Cummington Street, Boston, MA 02215, USA Activation of the Rel/NF-jB signal transduction pathway called the Rel homology domain, which has sequences has been associated with a varietyof animal and human required for DNA binding, dimerization, nuclear malignancies. However, among the Rel/NF-jB family localization, and inhibitor (IkB) binding. Sequences members, onlyc-Rel has been consistentlyshown to be C-terminal to the Rel homology domain in c-Rel, RelA, able to malignantlytransform cells in culture. In addition, and RelB contain transcriptional activation domains, c-rel has been activated bya retroviral promoter insertion and therefore dimers that contain these Rel/NF-kB in an avian B-cell lymphoma, and amplifications of REL members usually increase transcription of target genes. (human c-rel) are frequentlyseen in Hodgkin’s lympho- Although the Rel homology domain sequences of c-Rel mas and diffuse large B-cell lymphomas, and in some proteins across species are highly conserved, their follicular and mediastinal B-cell lymphomas. Phenotypic C-terminal transactivation domains are not. For exam- analysis of c-rel knockout mice demonstrates that c-Rel ple, the Rel homology domains of chicken and human has a normal role in B-cell proliferation and survival; c-Rel are approximately 85% identical, whereas their moreover, c-Rel nuclear activityis required for B-cell C-terminal transactivation domains are only about 10% development. -

Elimination of Plasmatocytes by Targeted Apoptosis Reveals Their Role in Multiple Aspects of the Drosophila Immune Response
Elimination of plasmatocytes by targeted apoptosis reveals their role in multiple aspects of the Drosophila immune response Bernard Charroux and Julien Royet1 Institut de Biologie du De´veloppement de Marseille Luminy, Unite´Mixte de Recherche 6216, Centre National de la Recherche Scientifique, Universite´dela Méditerrannée Aix-Marseille II, 13288 Marseille Cedex 9, France Communicated by Jules A. Hoffmann, Centre National de la Recherche Scientifique, Strasbourg, France, April 17, 2009 (received for review January 14, 2009) Drosophila hemocytes have strong phagocytic capacities and pro- generating plasmatocyte-depleted individuals and by analyzing duce antimicrobial peptides (AMPs). However, the precise role of their development and immune response. blood cells during immune responses and developmental processes has only been studied using indirect means. To overcome this Results limitation, we generated plasmatocyte-depleted flies by specifi- To obtain flies devoid of plasmatocytes, we decided to trigger cally overexpressing the proapoptotic protein Hid into plasmato- cell death in blood cells by overexpressing the proapoptotic cytes. Unexpectedly, these plasmatocyte-depleted animals have a protein Hid using the plasmatocyte (and crystal cells)–specific normal larval and pupal development and do not exhibit any hml(⌬)-Gal4 driver (20, 21). A UAS-EGFP transgene was used obvious defect after birth. Remarkably, plasmatocyte-depleted to precisely characterize the spatiotemporal expression of the adults show a strong susceptibility to infections by various micro- hml(⌬)-Gal4 driver. GFP is not expressed during embryogenesis, organisms, although activation of systemic AMP gene transcription even in late embryos in which Croquemort-positive blood cells via the Toll and immune deficiency (IMD) pathways is wild-type. are detected (Fig. -

The Role of Ubiquitination in NF-Κb Signaling During Virus Infection
viruses Review The Role of Ubiquitination in NF-κB Signaling during Virus Infection Kun Song and Shitao Li * Department of Microbiology and Immunology, Tulane University, New Orleans, LA 70112, USA; [email protected] * Correspondence: [email protected] Abstract: The nuclear factor κB (NF-κB) family are the master transcription factors that control cell proliferation, apoptosis, the expression of interferons and proinflammatory factors, and viral infection. During viral infection, host innate immune system senses viral products, such as viral nucleic acids, to activate innate defense pathways, including the NF-κB signaling axis, thereby inhibiting viral infection. In these NF-κB signaling pathways, diverse types of ubiquitination have been shown to participate in different steps of the signal cascades. Recent advances find that viruses also modulate the ubiquitination in NF-κB signaling pathways to activate viral gene expression or inhibit host NF-κB activation and inflammation, thereby facilitating viral infection. Understanding the role of ubiquitination in NF-κB signaling during viral infection will advance our knowledge of regulatory mechanisms of NF-κB signaling and pave the avenue for potential antiviral therapeutics. Thus, here we systematically review the ubiquitination in NF-κB signaling, delineate how viruses modulate the NF-κB signaling via ubiquitination and discuss the potential future directions. Keywords: NF-κB; polyubiquitination; linear ubiquitination; inflammation; host defense; viral infection Citation: Song, K.; Li, S. The Role of 1. Introduction Ubiquitination in NF-κB Signaling The nuclear factor κB (NF-κB) is a small family of five transcription factors, including during Virus Infection. Viruses 2021, RelA (also known as p65), RelB, c-Rel, p50 and p52 [1]. -

B Protein Relish Regulates the JNK-Mediated Immune Response in Drosophila
Downloaded from genesdev.cshlp.org on September 24, 2021 - Published by Cold Spring Harbor Laboratory Press Targeting of TAK1 by the NF-B protein Relish regulates the JNK-mediated immune response in Drosophila Jin Mo Park,1 Helen Brady,3 Maria Grazia Ruocco,1 Huaiyu Sun,2 DeeAnn Williams,2 Susan J. Lee,2 Tomohisa Kato Jr.,1 Normand Richards,3 Kyle Chan,3 Frank Mercurio,3 Michael Karin,1 and Steven A. Wasserman2,4 1Laboratory of Gene Regulation and Signal Transduction, Department of Pharmacology, School of Medicine, and 2Center for Molecular Genetics, Section of Cell and Developmental Biology, Division of Biology, University of California, San Diego, La Jolla, California 92093-0636, USA; 3Celgene Corporation, San Diego, California 92121, USA The molecular circuitry underlying innate immunity is constructed of multiple,evolutionarily conserved signaling modules with distinct regulatory targets. The MAP kinases and the IKK-NF-B molecules play important roles in the initiation of immune effector responses. We have found that the Drosophila NF-B protein Relish plays a crucial role in limiting the duration of JNK activation and output in response to Gram-negative infections. Relish activation is linked to proteasomal degradation of TAK1,the upstream MAP kinase kinase kinase required for JNK activation. Degradation of TAK1 leads to a rapid termination of JNK signaling,resulting in a transient JNK-dependent response that precede s the sustained induction of Relish-dependent innate immune loci. Because the IKK-NF-B module also negatively regulates JNK activation in mammals,thereby controlling inflammation-induced apoptosis,the regulatory cross-talk between the JNK and NF-B pathways appears to be broadly conserved. -

And C-Terminal Domains of Relb Are Required for Full Transactivation
MOLECULAR AND CELLULAR BIOLOGY, Mar. 1993, p. 1572-1582 Vol. 13, No. 3 0270-7306/93/031572-11$02.00/0 Copyright © 1993, American Society for Microbiology Both N- and C-Terminal Domains of RelB Are Required for Full Transactivation: Role of the N-Terminal Leucine Zipper-Like Motif PAWEL DOBRZANSKI, ROLF-PETER RYSECK, AND RODRIGO BRAVO* Department ofMolecular Biology, Bristol-Myers Squibb Pharmaceutical Research Institute, P.O. Box 4000, Princeton, New Jersey 08543-4000 Received 18 September 1992/Returned for modification 11 November 1992/Accepted 12 December 1992 RelB, a member of the Rel family of transcription factors, can stimulate promoter activity in the presence of p50-NF-KB or p5OB/p49-NF-KB in mammalian cells. Transcriptional activation analysis reveals that the N and C termini of RelB are required for full transactivation in the presence of p50-NF-KB. RelB/p50-NF-KB hybrid molecules containing the Rel homology domain of p50-NF-KB and the N and C termini of RelB have high transcriptional activity compared with wild-type p50-NF-KB. The N and C termini of RelB cooperate in transactivation in cis or trans configuration. Alterations in the structure of the leucine zipper-like motif present in the N terminus of RelB significantly decrease the transcriptional capacity of RelB and of different RelB/p50-NF-KB hybrid molecules. Serum stimulation of quiescent fibroblasts induces the regulated by the inhibitory factor cactus (28). The different expression of more than 100 genes, including several proto- homodimers and heterodimers formed between the members oncogenes encoding for transcriptional factors belonging to of the Rel family regulate gene expression in vivo (20, 22, 42) the Jun, Fos, and Rel families (2, 11, 13, 24, 34-36, 38). -

A Truncated TIR-NBS Protein TN10 Pairs with Two Clustered TIR-NBS-LRR Immune Receptors and Contributes to Plant Immunity in Arabidopsis
International Journal of Molecular Sciences Article A Truncated TIR-NBS Protein TN10 Pairs with Two Clustered TIR-NBS-LRR Immune Receptors and Contributes to Plant Immunity in Arabidopsis Yongming Chen 1, Guitao Zhong 1,2, Huiren Cai 1,2, Renjie Chen 1, Na Liu 1, Wei Wang 1,* and Dingzhong Tang 1,* 1 State Key Laboratory of Ecological Control of Fujian-Taiwan Crop Pests, Key Laboratory of Ministry of Education for Genetics, Breeding and Multiple Utilization of Crops, Plant Immunity Center, Fujian Agriculture and Forestry University, Fuzhou 350002, China; [email protected] (Y.C.); [email protected] (G.Z.); [email protected] (H.C.); [email protected] (R.C.); [email protected] (N.L.) 2 College of Life Science, Fujian Agriculture and Forestry University, Fuzhou 350002, China * Correspondence: [email protected] (W.W.); [email protected] (D.T.) Abstract: The encoding genes of plant intracellular nucleotide-binding site (NBS) and leucine-rich repeat (LRR) domain receptors (NLRs) often exist in the form of a gene cluster. Several recent studies demonstrated that the truncated Toll/interleukin-1 receptor-NBS (TIR-NBS) proteins play important roles in immunity. In this study, we identified a large TN gene cluster on Arabidopsis ecotype Col-0 chromosome 1, which included nine TN genes, TN4 to TN12. Interestingly, this cluster also contained two typical TIR-NBS-LRR genes: At1g72840 and At1g72860 (hereinafter referred to as TNL40 and Citation: Chen, Y.; Zhong, G.; TNL60, respectively), which formed head-to-head genomic arrangement with TN4 to TN12. However, Cai, H.; Chen, R.; Liu, N.; Wang, W.; the functions of these TN and TNL genes in this cluster are still unknown.