Open and Inclusive Collaboration in Science: a Framework”, OECD Science, Technology and Industry Working Papers, 2018/07, OECD Publishing, Paris
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Sweave User Manual
Sweave User Manual Friedrich Leisch and R-core June 30, 2017 1 Introduction Sweave provides a flexible framework for mixing text and R code for automatic document generation. A single source file contains both documentation text and R code, which are then woven into a final document containing • the documentation text together with • the R code and/or • the output of the code (text, graphs) This allows the re-generation of a report if the input data change and documents the code to reproduce the analysis in the same file that contains the report. The R code of the complete 1 analysis is embedded into a LATEX document using the noweb syntax (Ramsey, 1998) which is usually used for literate programming Knuth(1984). Hence, the full power of LATEX (for high- quality typesetting) and R (for data analysis) can be used simultaneously. See Leisch(2002) and references therein for more general thoughts on dynamic report generation and pointers to other systems. Sweave uses a modular concept using different drivers for the actual translations. Obviously different drivers are needed for different text markup languages (LATEX, HTML, . ). Several packages on CRAN provide support for other word processing systems (see Appendix A). 2 Noweb files noweb (Ramsey, 1998) is a simple literate-programming tool which allows combining program source code and the corresponding documentation into a single file. A noweb file is a simple text file which consists of a sequence of code and documentation segments, called chunks: Documentation chunks start with a line that has an at sign (@) as first character, followed by a space or newline character. -

Scholarly Communication and Data
Mooney, H. (2016). Scholarly communication and data. In L. M. Kellam & K. Thompson (Eds.), Databrarianship: The academic data librarian in theory and practice (pp. 195–218). Chicago: Association of College and Research Libraries. CHAPTER 13 Scholarly Communication and Data Hailey Mooney Introduction The Internet and digital data are strong forces shaping the modern world of schol- arly communication, the context within which academic librarians operate. Schol- arly communication entails the ways by which scholarly and research information are created, disseminated, evaluated, and preserved.1 Recognition of the broader forces at play in research and scholarship is imperative to keep ourselves from obsolescence and to simply function as effective librarians. The operation of the scholarly communication system is the “bedrock” of academic information litera- cy and forms the “sociocultural frame of reference” for understanding library re- search skills.2 The purpose of this chapter is to provide foundational knowledge for the data librarian by developing an understanding of the place of data within the current paradigm of networked digital scholarly communication. This includes defining the nature of data and data publications, examining the open science movement and its effects on data sharing, and delving into the challenges inherent to the wider integration of data into the scholarly communication system and the academic library. The Nature of Scientific Knowledge and Data The sociology of science provides a basis from which to understand the fundamen- tal underpinnings of the norms and values that govern the institution of science and the production of scientific knowledge. In basic information literacy instruc- tion, librarians teach the difference between popular and scholarly information. -

Exploring Controversies in the Circulation of Early SARS-Cov-2
Global Public Health An International Journal for Research, Policy and Practice ISSN: (Print) (Online) Journal homepage: https://www.tandfonline.com/loi/rgph20 Open science, COVID-19, and the news: Exploring controversies in the circulation of early SARS- CoV-2 genomic epidemiology research Stephen Molldrem, Mustafa I. Hussain & Anthony K J Smith To cite this article: Stephen Molldrem, Mustafa I. Hussain & Anthony K J Smith (2021): Open science, COVID-19, and the news: Exploring controversies in the circulation of early SARS-CoV-2 genomic epidemiology research, Global Public Health, DOI: 10.1080/17441692.2021.1896766 To link to this article: https://doi.org/10.1080/17441692.2021.1896766 © 2021 The Author(s). Published by Informa UK Limited, trading as Taylor & Francis Group Published online: 04 Mar 2021. Submit your article to this journal Article views: 30 View related articles View Crossmark data Full Terms & Conditions of access and use can be found at https://www.tandfonline.com/action/journalInformation?journalCode=rgph20 GLOBAL PUBLIC HEALTH https://doi.org/10.1080/17441692.2021.1896766 Open science, COVID-19, and the news: Exploring controversies in the circulation of early SARS-CoV-2 genomic epidemiology research Stephen Molldrem a, Mustafa I. Hussain b and Anthony K J Smith c aDepartment of Anthropology, University of California, Irvine, CA, USA; bDepartment of Informatics, University of California, Irvine, CA, USA; cCentre for Social Research in Health, UNSW Sydney, Sydney, Australia ABSTRACT ARTICLE HISTORY Some early English language news coverage of COVID-19 epidemiology Received 21 October 2020 focused on studies that examined how SARS-CoV-2 (the coronavirus Accepted 22 February 2021 that causes COVID-19) was evolving at the genetic level. -

Towards Open Science in Acoustics: Foundations and Best Practices
DAGA 2017 Kiel Towards Open Science in Acoustics: Foundations and Best Practices Sascha Spors1, Matthias Geier1 and Hagen Wierstorf2 1Institute of Communications Engineering, University of Rostock, Germany, Email: [email protected] 2Filmuniversit¨atBabelsberg KONRAD WOLF The Scientific Method myself Before introducing the open science approach in detail it my future self is worthwhile to review the foundations of the scientific my colleagues method. It may be referred to as `A method of procedure that has characterized natural science since the 17th cen- other researchers tury, consisting in systematic observation, measurement, all people in the world and experiment, and the formulation, testing, and modi- fication of hypotheses.' [1]. Science builds upon a set of science itself well accepted methods which are commonly believed to While research conducted for myself does not require any ensure above formulated principles. Besides these, the transparency in terms of underlying data, research per- reproducibility of results is one of the cornerstones of the formed for the sake of science itself should be as open as scientific method. It may be defined as `Reproducibil- possible to support scientific advancement. ity is the ability of an entire analysis of an experiment or study to be duplicated, either by the same researcher or by someone else working independently, whereas reproducing Open Science an experiment is called replicating it.' [2]. The irrepro- Open science follows the general demand for socializa- ducibility of a wide range of scientific results has drawn tion of knowledge. More specifically, it aims at making significant attention in the last decade [3, 4, 5, 6, 7, 8, 9]. -

ESS — Emacs Speaks Statistics
ESS — Emacs Speaks Statistics ESS version 5.14 The ESS Developers (A.J. Rossini, R.M. Heiberger, K. Hornik, M. Maechler, R.A. Sparapani, S.J. Eglen, S.P. Luque and H. Redestig) Current Documentation by The ESS Developers Copyright c 2002–2010 The ESS Developers Copyright c 1996–2001 A.J. Rossini Original Documentation by David M. Smith Copyright c 1992–1995 David M. Smith Permission is granted to make and distribute verbatim copies of this manual provided the copyright notice and this permission notice are preserved on all copies. Permission is granted to copy and distribute modified versions of this manual under the conditions for verbatim copying, provided that the entire resulting derived work is distributed under the terms of a permission notice identical to this one. Chapter 1: Introduction to ESS 1 1 Introduction to ESS The S family (S, Splus and R) and SAS statistical analysis packages provide sophisticated statistical and graphical routines for manipulating data. Emacs Speaks Statistics (ESS) is based on the merger of two pre-cursors, S-mode and SAS-mode, which provided support for the S family and SAS respectively. Later on, Stata-mode was also incorporated. ESS provides a common, generic, and useful interface, through emacs, to many statistical packages. It currently supports the S family, SAS, BUGS/JAGS, Stata and XLisp-Stat with the level of support roughly in that order. A bit of notation before we begin. emacs refers to both GNU Emacs by the Free Software Foundation, as well as XEmacs by the XEmacs Project. The emacs major mode ESS[language], where language can take values such as S, SAS, or XLS. -

Article (Published Version)
Article Open Science in Switzerland: Opportunities and Challenges PFISTER, Roger, et al. Abstract Open Science stands for a new approach to the scientific process, based on cooperative work and new ways of making knowledge available. It is thus an umbrella term for various movements aiming to remove the barriers to sharing any kind of output, resources, methods or tools, at any stage of the research process.Here, we focus on the open access to scientific literature and to data because of their particularly high relevance to the scientific community in Switzerland, at which this factsheet is primarily addressed. Both Open Access and Open Data are important science policy topics in different parts of the world, but the developments in Europe are most pertinent for Switzerland. This factsheet therefore presents the issues at stake in the on-going discussion in Europe and Switzerland, points out opportunities and addresses challenges. The recommendations are guided by the key consideration to shape Open Access and Open Data so that they foster scientific progress and benefit society. Reference PFISTER, Roger, et al. Open Science in Switzerland: Opportunities and Challenges. Swiss Academies Factsheets, 2019, vol. 14, no. 2 DOI : 10.5281/zenodo.3248929 Available at: http://archive-ouverte.unige.ch/unige:122751 Disclaimer: layout of this document may differ from the published version. 1 / 1 Vol. 14, No. 2, 2019 www.swiss-academies.ch Open Science in Switzerland: Opportunities and Challenges Open Science stands for a new approach to the scientific process, based on cooperative work and new ways of making knowledge available. It is thus an umbrella term for various movements aiming to remove the barriers to sharing any kind of output, resources, methods or tools, at any stage of the research process (Figure 1).1 Here, we focus on the open access to scientific literature and to data because of their particularly high relevance to the scientific community in Switzerland, at which this factsheet is primarily addressed. -

Open Science, the Challenge of Transparency 30
BERNARD RENTIER OPEN SCIENCE, THE CHALLENGE OF TR ANSPARENCY Preface by Philippe Busquin ACADÉMIE ROYALE DE BELGIQUE Collection L’ACADÉMIE EN POCHE OPEN SCIENCE, THE CHALLENGE OF TRANSPARENCY BERNARD RENTIER OPEN SCIENCE, THE CHALLENGE OF TRANSPARENCY Preface by Philippe Busquin ACADÉMIE ROYALE DE BELGIQUE Collection L’ACADÉMIE EN POCHE- With the support of Académie royale de Belgique Credits: rue Ducale, 1 © Translated from French by the author, 1 000 Brussels, Belgium Bernard Rentier www.academie-editions.be www.academieroyale.be Graphic design: Loredana Buscemi, Series Pocket Book Academy Académie royale de Belgique Under the academic responsibility of Didier Viviers ISBN 978-2-8031-0667-7 Volume 118-EN Dépôt légal : 2019/0092/3 © 2019, Académie royale de Belgique To Alma Swan, my friendly mentor in Open Access activism. Preface Writing the preface to this book published in L’Académie en Poche and jointly, for the first time, in a free open access electronic version, is a pleasure and an honour. Bernard Rentier, beyond his academic quali- ties and his rectoral responsibilities at the University of Liège, is a tireless and committed activist who shares with us his fight for Open Science. In this respect, it is entirely in line with the approach advocated by the European Commission and which Carlos Moedas, the current European Commissioner for Research, has also made his main focus. The 2016 European Union Report on Science, Research and Innovation is subtitled: “Contri- bution to Open Innovation, Open Science and Open to the World Agenda”, indicating the EU’s commitment to developing high quality science and positioning itself as a global leader in Open 9 OPEN SCIENCE, THE CHALLENGE OF TRANSPARENCY Science. -

Species' Traits and Phylogenetic
Northern Michigan University NMU Commons All NMU Master's Theses Student Works 8-2018 CLIMATE DRIVEN RANGE SHIFTS OF NORTH AMERICAN SMALL MAMMALS: SPECIES’ TRAITS AND PHYLOGENETIC INFLUENCES Katie Nehiba [email protected] Follow this and additional works at: https://commons.nmu.edu/theses Part of the Ecology and Evolutionary Biology Commons Recommended Citation Nehiba, Katie, "CLIMATE DRIVEN RANGE SHIFTS OF NORTH AMERICAN SMALL MAMMALS: SPECIES’ TRAITS AND PHYLOGENETIC INFLUENCES" (2018). All NMU Master's Theses. 557. https://commons.nmu.edu/theses/557 This Open Access is brought to you for free and open access by the Student Works at NMU Commons. It has been accepted for inclusion in All NMU Master's Theses by an authorized administrator of NMU Commons. For more information, please contact [email protected],[email protected]. CLIMATE DRIVEN RANGE SHIFTS OF NORTH AMERICAN SMALL MAMMALS: SPECIES’ TRAITS AND PHYLOGENETIC INFLUENCES By Katie R. Nehiba THESIS Submitted to Northern Michigan University In partial fulfillment of the requirements For the degree of MASTER OF SCIENCE Office of Graduate Education and Research July 2018 ABSTRACT By Katie R. Nehiba Current anthropogenically-driven climate change is accelerating at an unprecedented rate. In response, species’ ranges may shift, tracking optimal climatic conditions. Species-specific differences may produce predictable differences in the extent of range shifts. I evaluated if patterns of predicted responses to climate change were strongly related to species’ taxonomic identities and/or ecological characteristics of species’ niches, elevation and precipitation. I evaluated differences in predicted range shifts in well-sampled small mammals that are restricted to North America: kangaroo rats, voles, chipmunks, and ground squirrels. -

Reproducible Research.. Using Sweave, Knitr and Pandoc
Reproducible Research.. Using Sweave, Knitr and Pandoc Aedín Culhane [email protected] Nov 20th 2012 My R Course Website http://bcb.dfci.harvard.edu/~aedin/ My HSPH homepage http://www.hsph.harvard.edu/research/aedin-culhane/ When issues of reproducibility arise • ``Remember that microarray analysis you did six months ago? We ran a few more arrays. Can you add them to the project and repeat the same analysis?'' • ``The statistical analyst who looked at the data I generated previously is no longer available. Can you get someone else to analyze my new data set using the same methods (and thus producing a report I can expect to understand)?'' • ``Please write/edit the methods sections for the abstract/paper/grant proposal I am submitting based on the analysis you did several months ago.'' From Keith Baggerly • Selected articles published in Nature Genetics between January 2005 and December 2006 that had used profiling with microarrays • Of the 56 items retrieved electronically, 20 articles were considered potentially eligible for the project • The four teams were from – University of Alabama at Birmingham (UAB) – Stanford/Dana-Farber (SD) – London (L) and Ioannina/Trento (IT) • Each team was comprised of 3-6 scientists who worked together to evaluate each article. Results • Result could be reproduced n=2 • Reproduced with discrepancy n=6 • Could not be reproduced n=10 – No data n=4 (no data n=2, subset n=1, no reporter data n=1) – Confusion over matching of data to analysis (n=2) – Specialized software required and not available (n=1)m – Raw data available but could not be processed n=2 Reproducibility of Analysis Ioannidis JP, Allison DB, Ball CA, Coulibaly I, Cui X, Culhane AC, et al, (2009) Repeatability of published microarray gene expression Analyses. -

Ethical Questions in Open Science
Ethical Questions in Open Science Ariel Deardorff, MLIS Data Services Librarian UCSF Library About Me Data Services Librarian Open Science Researcher Situated in the Library’s Data Science Team Today’s Talk ● Explore Open Science ● COVID-19 & Open Access ● COVID-19 & Open Data ● Unpack Ethical Issues What is open science? (slides from Open Science 101 co-developed with Anneliese Taylor) “Closed” science workflow Paywall “Open science is the movement to make scientific research, data, and dissemination accessible to all levels of an inquiring society.” - FOSTER Open Science Open science workflow Open Access Open Data Open Methods Open Protocols Open Science = Open Source Open Code Open Educational Resources Open Pedagogy Why practice open science? Requirements - this is an evolving area! Funders: The NIH has had an open access mandate for publications since 2008 and a genomic data sharing mandate since 2014. Will likely have a comprehensive data sharing mandate by the end of 2021. Institutions: The University of California also has an open access mandate Journals: Several journals (PLOS, Nature, Science) now require open data to publish. Get Credit for your Work! You work hard on your data, code, and methods and they should be recognized as citable research outputs that go on your CV Making your work open leads to more citations for your articles: - Piwowar et al. found that articles published as open access receive 18% more citations than average - A study from Colavizza et al. showed that studies that provide access to underlying data are -
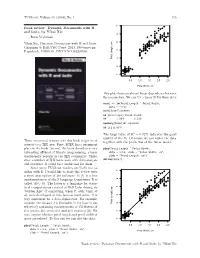
Dynamic Documents with R and Knitr, by Yihui Xie 116 Tugboat, Volume 35 (2014), No
TUGboat, Volume 35 (2014), No. 1 115 ● ● ● Book review: Dynamic Documents with R ● ● ● and knitr, by Yihui Xie ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● Boris Veytsman ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● Yihui Xie, Dynamic Documents with R and knitr. ● ● ● ● ● ● ● ● ● ● ● ● ● ● Chapman & Hall/CRC Press, 2013, 190+xxvi pp. ● ● ● ● ● ● ● ● ● US$ ISBN ● Paperback, 59.95. 978-1482203530. ● ● ● ● Petal Length, cm Petal ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● 1 2 3 4 5 6 7 0.5 1.0 1.5 2.0 2.5 Petal Width, cm This plot shows an almost linear dependence between the parameters. We can try a linear fit for these data: model <- lm(Petal.Length ˜ Petal.Width, data = iris) model$coefficients ## (Intercept) Petal.Width ## 1.084 2.230 summary(model)$r.squared ## [1] 0.9271 The large value of R2 = 0.9271 indicates the good quality of the fit. Of course we can replot the data There are several reasons why this book might be of together with the prediction of the linear model: interest to a TEX user. First, LATEX has a prominent place in the book. Second, the book describes a very plot(Petal.Length ˜ Petal.Width, interesting offshoot of literate programming, a topic data = iris, xlab = "Petal Width, cm", traditionally popular in the TEX community. Third, ylab = "Petal Length, cm") abline(model) since a number of TEX users work with data analysis and statistics, R could be a useful tool for them. Since some TUGboat readers are likely not fa- ● ● ● miliar with R, I would like to start this review with ● ● a short description of the software. R [1] is a free ● ● ● ● ● ● ● ● ● ● ● implementation of the S language (sometimes R is ● ● ● ● ● ● ● ● ● ● ● ● ● ● called GNU S). -

LYX and Knitr
knitr.1 LYX and knitr The knitr package allows one to embed R code within LYX and LATEX documents. When a document is compiled into a PDF, LYX/LATEX connects to R to run the R code and the code/output is automatically put into the PDF. In addition to this being a convenient way to use both LYX/LATEX with R, it also provides an important component to the reproducibility of research (RR). For example, one can include the code for a data analysis de- scribed in a paper. This ensures that there would be no “copying and pasting errors” and also provide readers of the paper an im- mediate way to reproduce the research. RR continues to become more important and fortunately more tools are being developed to make it possible. Below are some discussions on the topic: • AMSTAT News column on RR at http://magazine. amstat.org/blog/2011/01/01/scipolicyjan11. • CRAN task view for RR and R at http://cran.r-project. org/web/views/ReproducibleResearch.html. • Yihui Xie: Author of knitr – First and second editions of his Dynamic Documents with R and knitr book. Note that this book was typed in LYX. – Website for knitr at http://yihui.name/knitr The Sweave environment is another way to include R code inside of LYX/LATEX. This was developed prior to knitr, but it is more difficult to use. The purpose of this section is to examine the main compo- nents of knitr so that you will be able to complete the rest of the semester using LYX and knitr together for all assign- ments in our course! Also, a very important purpose is to give you the tools needed to complete all assignments in other R- based courses by using knitr and LYX together! The files used knitr.2 here are intro_example_cereal.lyx, intro_example_cereal.pdf, cereal.csv, ExternalCode.R, FirstBeamer-knitr.zip, JSM2015.zip, and RMarkdown.zip.