Phd Dissertation
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

How Influenza Virus Uses Host Cell Pathways During Uncoating
cells Review How Influenza Virus Uses Host Cell Pathways during Uncoating Etori Aguiar Moreira 1 , Yohei Yamauchi 2 and Patrick Matthias 1,3,* 1 Friedrich Miescher Institute for Biomedical Research, 4058 Basel, Switzerland; [email protected] 2 Faculty of Life Sciences, School of Cellular and Molecular Medicine, University of Bristol, Bristol BS8 1TD, UK; [email protected] 3 Faculty of Sciences, University of Basel, 4031 Basel, Switzerland * Correspondence: [email protected] Abstract: Influenza is a zoonotic respiratory disease of major public health interest due to its pan- demic potential, and a threat to animals and the human population. The influenza A virus genome consists of eight single-stranded RNA segments sequestered within a protein capsid and a lipid bilayer envelope. During host cell entry, cellular cues contribute to viral conformational changes that promote critical events such as fusion with late endosomes, capsid uncoating and viral genome release into the cytosol. In this focused review, we concisely describe the virus infection cycle and highlight the recent findings of host cell pathways and cytosolic proteins that assist influenza uncoating during host cell entry. Keywords: influenza; capsid uncoating; HDAC6; ubiquitin; EPS8; TNPO1; pandemic; M1; virus– host interaction Citation: Moreira, E.A.; Yamauchi, Y.; Matthias, P. How Influenza Virus Uses Host Cell Pathways during 1. Introduction Uncoating. Cells 2021, 10, 1722. Viruses are microscopic parasites that, unable to self-replicate, subvert a host cell https://doi.org/10.3390/ for their replication and propagation. Despite their apparent simplicity, they can cause cells10071722 severe diseases and even pose pandemic threats [1–3]. -

Hutchinson, EC, & Yamauchi, Y
Hutchinson, E. C., & Yamauchi, Y. (2018). Understanding Influenza. In Influenza Virus: Methods and Protocols (pp. 1-21). (Methods in Molecular Biology; Vol. 1836). Humana Press. https://doi.org/10.1007/978-1-4939-8678-1_1 Peer reviewed version Link to published version (if available): 10.1007/978-1-4939-8678-1_1 Link to publication record in Explore Bristol Research PDF-document This is the author accepted manuscript (AAM). The final published version (version of record) is available online via Springer Nature at https://link.springer.com/protocol/10.1007%2F978-1-4939-8678-1_1. Please refer to any applicable terms of use of the publisher. University of Bristol - Explore Bristol Research General rights This document is made available in accordance with publisher policies. Please cite only the published version using the reference above. Full terms of use are available: http://www.bristol.ac.uk/red/research-policy/pure/user-guides/ebr-terms/ Understanding Influenza Edward C. Hutchinson1* and Yohei Yamauchi2* 1MRC-University of Glasgow Centre for Virus Research; 2School of Cellular and Molecular Medicine, University of Bristol. *Corresponding authors: [email protected], [email protected] Running Head: Understanding Influenza Abstract Influenza, a serious illness of humans and domesticated animals, has been studied intensively for many years. It therefore provides an example of how much we can learn from detailed studies of an infectious disease, and of how even the most intensive scientific research leaves further questions to answer. This introduction is written for researchers who have become interested in one of these unanswered questions, but who may not have previously worked on influenza. -

University of Groningen Molecular Insights Into Viral Respiratory Infections Cong, Ying-Ying
University of Groningen Molecular insights into viral respiratory infections Cong, Ying-Ying IMPORTANT NOTE: You are advised to consult the publisher's version (publisher's PDF) if you wish to cite from it. Please check the document version below. Document Version Publisher's PDF, also known as Version of record Publication date: 2019 Link to publication in University of Groningen/UMCG research database Citation for published version (APA): Cong, Y-Y. (2019). Molecular insights into viral respiratory infections. University of Groningen. Copyright Other than for strictly personal use, it is not permitted to download or to forward/distribute the text or part of it without the consent of the author(s) and/or copyright holder(s), unless the work is under an open content license (like Creative Commons). Take-down policy If you believe that this document breaches copyright please contact us providing details, and we will remove access to the work immediately and investigate your claim. Downloaded from the University of Groningen/UMCG research database (Pure): http://www.rug.nl/research/portal. For technical reasons the number of authors shown on this cover page is limited to 10 maximum. Download date: 25-09-2021 CHAPTER I General Introduction Chapter I The structure of the respiratory tract facilitates gas exchange between the exterior environment and interior milieu of the host, while it is a susceptible target and feasible gateway for diverse pathogens. Pandemics of severe acute respiratory infections have been serious threats to global health, causing significant morbidity and mortality. In particular, influenza viruses and coronaviruses (CoV), including MERS-CoV and SARS-CoV, have caused numerous outbreaks of viral pneumonia worldwide with different impacts. -

Single Mosquito Metatranscriptomics Identifies Vectors, Emerging Pathogens and Reservoirs in One Assay
TOOLS AND RESOURCES Single mosquito metatranscriptomics identifies vectors, emerging pathogens and reservoirs in one assay Joshua Batson1†, Gytis Dudas2†, Eric Haas-Stapleton3†, Amy L Kistler1†*, Lucy M Li1†, Phoenix Logan1†, Kalani Ratnasiri4†, Hanna Retallack5† 1Chan Zuckerberg Biohub, San Francisco, United States; 2Gothenburg Global Biodiversity Centre, Gothenburg, Sweden; 3Alameda County Mosquito Abatement District, Hayward, United States; 4Program in Immunology, Stanford University School of Medicine, Stanford, United States; 5Department of Biochemistry and Biophysics, University of California San Francisco, San Francisco, United States Abstract Mosquitoes are major infectious disease-carrying vectors. Assessment of current and future risks associated with the mosquito population requires knowledge of the full repertoire of pathogens they carry, including novel viruses, as well as their blood meal sources. Unbiased metatranscriptomic sequencing of individual mosquitoes offers a straightforward, rapid, and quantitative means to acquire this information. Here, we profile 148 diverse wild-caught mosquitoes collected in California and detect sequences from eukaryotes, prokaryotes, 24 known and 46 novel viral species. Importantly, sequencing individuals greatly enhanced the value of the biological information obtained. It allowed us to (a) speciate host mosquito, (b) compute the prevalence of each microbe and recognize a high frequency of viral co-infections, (c) associate animal pathogens with specific blood meal sources, and (d) apply simple co-occurrence methods to recover previously undetected components of highly prevalent segmented viruses. In the context *For correspondence: of emerging diseases, where knowledge about vectors, pathogens, and reservoirs is lacking, the [email protected] approaches described here can provide actionable information for public health surveillance and †These authors contributed intervention decisions. -

Structures of Human-Infecting Thogotovirus Fusogens Support a Common Ancestor with Insect Baculovirus
Structures of human-infecting Thogotovirus fusogens PNAS PLUS support a common ancestor with insect baculovirus Ruchao Penga,b, Shuijun Zhanga,1, Yingzi Cuia,b, Yi Shia,b,c,d, George F. Gaoa,b,c,d,e,2, and Jianxun Qia,b,2 aChinese Academy of Sciences Key Laboratory of Pathogenic Microbiology and Immunology, Institute of Microbiology, Chinese Academy of Sciences, Beijing 100101, China; bSchool of Life Sciences, University of Chinese Academy of Sciences, Beijing 101408, China; cShenzhen Key Laboratory of Pathogen and Immunity, Shenzhen Third People’s Hospital, Shenzhen 518112, China; dCenter for Influenza Research and Early-Warning, Chinese Academy of Sciences, Beijing 100101, China; and eNational Institute for Viral Disease Control and Prevention, Chinese Center for Disease Control and Prevention, Beijing 102206, China Edited by Michael G. Rossmann, Purdue University, West Lafayette, IN, and approved September 12, 2017 (received for review April 12, 2017) Thogotoviruses are emerging tick-borne zoonotic orthomyxoviruses ilarity with the glycoproteins of influenza viruses or isavirus, infecting both humans and domestic animals with severe clinical indicating a distinct mechanism for entering host cells. The closest consequences. These viruses utilize a single-envelope glycoprotein orthomyxovirus relative of thogotovirus is quaranjavirus, whose (Gp) to facilitate their entry into host cells. Here, we present the Gp envelope Gp shares a sequence identity of ∼26% with thogotovirus structures of Thogoto and Dhori viruses, both of which are members Gps (11). Thus far the receptor of thogotoviruses has not been of the Thogotovirus genus in the family Orthomyxoviridae.These identified, and their entry pathway is unclear. The only evidence is structures, determined in the postfusion conformation, identified that the BOUV viral particles could be observed in the endosomal them as class III viral fusion proteins. -

Unprecedented Genomic Diversity of RNA Viruses In
RESEARCH ARTICLE elifesciences.org Unprecedented genomic diversity of RNA viruses in arthropods reveals the ancestry of negative-sense RNA viruses Ci-Xiu Li1,2†, Mang Shi1,2,3†, Jun-Hua Tian4†, Xian-Dan Lin5†, Yan-Jun Kang1,2†, Liang-Jun Chen1,2, Xin-Cheng Qin1,2, Jianguo Xu1,2, Edward C Holmes1,3, Yong-Zhen Zhang1,2* 1State Key Laboratory for Infectious Disease Prevention and Control, National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention, Beijing, China; 2Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, Hangzhou, China; 3Marie Bashir Institute for Infectious Diseases and Biosecurity, Charles Perkins Centre, School of Biological Sciences and Sydney Medical School, The University of Sydney, Sydney, Australia; 4Wuhan Center for Disease Control and Prevention, Wuhan, China; 5Wenzhou Center for Disease Control and Prevention, Wenzhou, China Abstract Although arthropods are important viral vectors, the biodiversity of arthropod viruses, as well as the role that arthropods have played in viral origins and evolution, is unclear. Through RNA sequencing of 70 arthropod species we discovered 112 novel viruses that appear to be ancestral to much of the documented genetic diversity of negative-sense RNA viruses, a number of which are also present as endogenous genomic copies. With this greatly enriched diversity we revealed that arthropods contain viruses that fall basal to major virus groups, including the vertebrate-specific *For correspondence: arenaviruses, filoviruses, hantaviruses, influenza viruses, lyssaviruses, and paramyxoviruses. We [email protected] similarly documented a remarkable diversity of genome structures in arthropod viruses, including †These authors contributed a putative circular form, that sheds new light on the evolution of genome organization. -

A Short Note on Orthomyxoviridae Y
Short Communication 1 A Short note on Orthomyxoviridae Y. Sai Sampath Kumar Andhra Loyola College, Vijayawada, India. Abstract disorder -A, − B and -C viruses, are enclosed polymer viruses that cause higher tract infections characterised Orthomyxoviridae (ὀρθός, orthós, Greek for by fever, chills, headache, generalized muscular aching, “straight”; μύξα, mýxa, Greek for “mucus”) could be a and loss of appetency (Webster et al. (1985)). The family family of negative-sense polymer viruses. It includes Orthomyxoviridae contains the genera Influenzavirus seven genera: Alphainfluenzavirus, Betainfluenzavirus, A, Influenzavirus B, Influenzavirus C, Thogotovirus, Deltainfluenzavirus, Gammainfluenzavirus, Isavirus, Quaranjavirus, and Isavirus. The name of the family Thogotovirus, and Quaranjavirus. The orthomyxoviruses comes from the Greek myxa, which means secretion, and (influenza viruses) represent the genus myxovirus, Keywordsorthos, which means correct or right. that consists of 3 sorts (species): A, B, and C. These viruses cause respiratory disorder, associate degree Orthomyxoviridae; Alphainfluenzavirus; Betainfluenzavirus ; acute disease with outstanding general symptoms. Deltainfluenzavirus The orthomyxoviridae family, containing respiratory Correspondence to: Citation: Sai Sampath Y. (2021). Cluster Analysis of Rabies Virus-Host (Homosapiens) Network to Determine Various Viral Y. Sai Sampath Kumar, Infections. EJBI. 17(6):01 Andhra Loyola College, DOI: 10.24105/ejbi.2020.17.6.01 Vijayawada, Received: June 01, 2021 India, Accepted: June 15, 2021 E-mail: [email protected] Published: June 22, 2021 1. Introduction Because the infectious agent order carries the blueprint for manufacturing new viruses, virologists think about it the foremost vital The myxovirus order contains eight segments of fibre negative-sense characteristic for classification. Respiratory disorder could be a fiber, polymer (ribonucleic acid), associate degreed an endogenous polymer helically formed, polymer virus of the myxovirus family. -

Neotropical Notebooks Please Include During a Visit on 9 April 1994 (Pyle Et Al
COTINGA 1 Neotropical Notebook Neotropical Notebook These recent reports generally refer to new or Chiriqui, during fieldwork between 1987 and 1991, second country records, rediscoveries, notable representing a disjunct population from that of Mexico range extensions, and new localities for threat to north-western Costa Rica (Olson 1993). Red- ened or poorly known species. These have been throated Caracara Daptrius americanus has been collated from a variety of published and unpub rediscovered in western Panama, with several seen and lished sources, and therefore some records will be heard on 26 August 1993 around the indian village of unconfirmed. We urge that, if they have not al Teribe (Toucan 19[9]: 5). ready done so, contributors provide full details to the relevant national organisations. COLOMBIA Recent expeditions and increasing interest in this coun BELIZE try has produced a wealth of new information, including There are five new records for the country as follows: a 12 new country records. A Cambridge–RHBNC expedi light phase Pomarine Jaeger Stercorarius pomarinus tion to Serranía de Naquén, Amazonas, in July–August seen by the fisheries pier, Belize City, 1 May 1992; 1992 found 4 new country records as follows: Rusty several Fulvous Whistling-Duck Dendrocygna bicolor Tinamou Crypturellus brevirostris observed at an ant- seen at Cox Lagoon in November 1986, up to 20 at swarm at Caño Ima, 12 August; Brown-banded Crooked Tree in March 1988, and again on 3 May 1992; Puffbird Notharchus tricolor observed in riverside a Chuck-will’s Widow Caprimulgus carolinensis col trees between Mahimachi and Caño Colorado [no date]; lected at San Ignacio, Cayo District, 13 October 1991; and a male Guianan Gnatcatcher Polioptila guianensis Spectacled Foliage-gleaner Anabacerthia observed at close range in a mixed flock at Caño Rico, 2 variegaticeps recently recorded on an expedition to the August (Amazon 1992). -

Downloaded from NCBI on Mar 27, 2019.) 636 Default Parameters Were Used, Except the E-Value Cutoff Was Set to 1E-2
bioRxiv preprint doi: https://doi.org/10.1101/2020.02.10.942854; this version posted February 13, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. bioRxiv PREPRINT 1 Single MOSQUITO 2 METATRANSCRIPTOMICS RECOVERS 3 MOSQUITO species, BLOOD MEAL 4 SOURces, AND MICROBIAL CARgo, 5 INCLUDING VIRAL DARK MATTER 1† 3† 2† 1*† 6 Joshua Batson , Gytis Dudas , Eric Haas-Stapleton , Amy L. Kistler , Lucy M. 1† 1† 1,4† 5† 7 Li , Phoenix Logan , Kalani Ratnasiri , Hanna Retallack *For CORRespondence: Chan ZuckERBERG Biohub, 499 ILLINOIS St, San FrANCISCO CA 94158; Alameda County [email protected] (AK) 8 1 2 Mosquito Abatement District, 23187 Connecticut St., Hayward, CA 94545; 3GothenburG † 9 These AUTHORS CONTRIBUTED EQUALLY Global Biodiversity Centre, Carl SkOTTSBERGS GATA 22B, 413 19, Gothenburg, Sweden; TO THIS WORK 10 PrOGRAM IN IMMUNOLOGY, StanforD University School OF Medicine, StanforD CA 94305; 11 4 Department OF Biochemistry AND Biophysics, University OF California San Francisco, San 12 5 FrANCISCO CA 94158 13 14 15 AbstrACT Mosquitoes ARE A DISEASE VECTOR WITH A COMPLEX ECOLOGY INVOLVING INTERACTIONS BETWEEN 16 TRANSMISSIBLE pathogens, ENDOGENOUS MICRobiota, AND HUMAN AND ANIMAL BLOOD MEAL SOURces. 17 Unbiased METATRANSCRIPTOMIC SEQUENCING OF INDIVIDUAL MOSQUITOES OffERS A STRAIGHTFORWARD AND 18 RAPID WAY TO CHARACTERIZE THESE dynamics. Here, WE PROfiLE 148 DIVERSE wild-caught MOSQUITOES 19 COLLECTED IN California, DETECTING SEQUENCES FROM eukaryotes, PRokaryotes, AND OVER 70 KNOWN AND 20 NOVEL VIRAL species. -
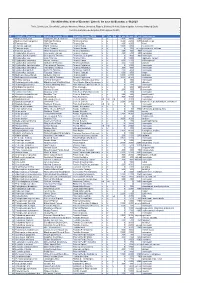
Ecuador Simple List Version 2020 Clements
Checklist of the birds of Ecuador / Lista de las aves del Ecuador, v. 08.2020 Freile, Brinkhuizen, Greenfield, Lysinger, Navarrete, Nilsson, Olmstead, Ridgely, Sánchez-Nivicela, Solano-Ugalde, Athanas, Ahlman & Boyla Comité Ecuatoriano de Registros Ornitológicos (CERO) ID Scientific_Clements_2019 English_Clements_2019 Español Ecuador CERO EC Con Gal Alt_min Alt_max Alt_ext Subespecies 1 Nothocercus julius Tawny-breasted Tinamou Tinamú Pechileonado x x 2300 3400 2100 monotypic 2 Nothocercus bonapartei Highland Tinamou Tinamú Serrano x x 1600 2200 3075 plumbeiceps 3 Tinamus tao Gray Tinamou Tinamú Gris x x 400 1600 kleei 4 Tinamus osgoodi Black Tinamou Tinamú Negro x x 1000 1400 hershkovitzi 5 Tinamus major Great Tinamou Tinamú Grande x x 0 700 1200, 1350 peruvianus, latifrons 6 Tinamus guttatus White-throated Tinamou Tinamú Goliblanco x x 200 400 900 monotypic 7 Crypturellus cinereus Cinereous Tinamou Tinamú Cinéreo x x 200 600 900 monotypic 8 Crypturellus berlepschi Berlepsch's Tinamou Tinamú de Berlepsch x x 0 400 900 monotypic 9 Crypturellus soui Little Tinamou Tinamú Chico x x 0 1200 nigriceps, harterti 10 Crypturellus obsoletus Brown Tinamou Tinamú Pardo x x 500 1100 chirimotanus? 11 Crypturellus undulatus Undulated Tinamou Tinamú Ondulado x x 200 600 yapura 12 Crypturellus transfasciatus Pale-browed Tinamou Tinamú Cejiblanco x x 0 1600 monotypic 13 Crypturellus variegatus Variegated Tinamou Tinamú Abigarrado x x 200 400 monotypic 14 Crypturellus bartletti Bartlett's Tinamou Tinamú de Bartlett x x 200 400 monotypic 15 Crypturellus -

Avian Influenza Infections in Nonmigrant Land Birds in Andean Peru
DOI: 10.7589/2011-02-052 Journal of Wildlife Diseases, 48(4), 2012, pp. 910–917 # Wildlife Disease Association 2012 View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by UDORA - University of Derby Online Research Archive AVIAN INFLUENZA INFECTIONS IN NONMIGRANT LAND BIRDS IN ANDEAN PERU Richard A. J. Williams,1,2,6 Karen Segovia-Hinostroza,3 Bruno M. Ghersi,3,4 Victor Gonzaga,4 A. Townsend Peterson,1 and Joel M. Montgomery4,5 1 Biodiversity Institute, 1345 Jayhawk Blvd., University of Kansas, Lawrence, Kansas 66045, USA 2 Departamento de Zoologı´a y Antropologı´aFı´sica, Universidad Complutense de Madrid, C/Jose´ Antonio Novais, 2 Ciudad Universitaria, 28040 Madrid, Spain 3 Facultad de Mecicina Veterinaria de Universidad Nacional Mayor de San Marcos, Av. Circunvalacio´n Cdra. 28 San Borja, Lima, Peru 4 United States Naval Medical Research Center Detachment, Unit 6, Av. Venezuela Cdra. 36, Callao 2, Lima, Peru 5 Current address: International Emerging Infections Program, GDDER-KenyaCDC-KenyaMbagathi Rd, Off Mbagathi Way, PO Box 606, Village Market 00621, Nairobi, Kenya 6 Corresponding author (email: [email protected]) ABSTRACT: As part of ongoing surveillance for avian influenza viruses (AIV) in Peruvian birds, in June 2008, we sampled 600 land birds of 177 species, using real-time reverse-transcription PCR. We addressed the assumption that AIV prevalence is low or nil among land birds, a hypothesis that was not supported by the results—rather, we found AIV infections at relatively high prevalences in birds of the orders Apodiformes (hummingbirds) and Passeriformes (songbirds). -

Characterization of Three Novel Viruses from the Families
viruses Article Characterization of Three Novel Viruses from the Families Nyamiviridae, Orthomyxoviridae, and Peribunyaviridae, Isolated from Dead Birds Collected during West Nile Virus Surveillance in Harris County, Texas Peter J. Walker 1 , Robert B. Tesh 2,3,4,5,6, Hilda Guzman 2,5,6, Vsevolod L. Popov 2,4,5,6, 2,5,6, 7 8 Amelia P.A. Travassos da Rosa y, Martin Reyna , Marcio R.T. Nunes , William Marciel de Souza 8,9 , Maria A. Contreras-Gutierrez 10, Sandro Patroca 8, Jeremy Vela 7, Vence Salvato 7, Rudy Bueno 7, Steven G. Widen 11 , Thomas G. Wood 11 and Nikos Vasilakis 2,3,4,5,6,* 1 School of Biological Sciences, The University of Queensland, St Lucia QLD 4072, Australia; [email protected] 2 Department of Pathology, University of Texas Medical Branch, 301 University Blvd, Galveston, TX 77555, USA; [email protected] (R.B.T.); [email protected] (H.G.); [email protected] (V.L.P.) 3 Center for Biodefense and Emerging Infectious Diseases, University of Texas Medical Branch, 301 University Blvd, Galveston, TX 77555, USA 4 Center for Tropical Diseases, University of Texas Medical Branch, 301 University Blvd, Galveston, TX 77555, USA 5 Institute for Human Infection and Immunity, University of Texas Medical Branch, 301 University Blvd, Galveston, TX 77555, USA 6 World Reference Center for Emerging Viruses and Arboviruses, University of Texas Medical Branch, 301 University Blvd, Galveston, TX 77555, USA 7 Mosquito and Vector Control Division, Harris County Public Health and Environmental Services, 3330 Old Spanish Trail, Houston, TX 77021,