Cooperative Secretions Facilitate Host Range Expansion in Bacteria
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

(Batch Learning Self-Organizing Maps), to the Microbiome Analysis of Ticks
Title A novel approach, based on BLSOMs (Batch Learning Self-Organizing Maps), to the microbiome analysis of ticks Nakao, Ryo; Abe, Takashi; Nijhof, Ard M; Yamamoto, Seigo; Jongejan, Frans; Ikemura, Toshimichi; Sugimoto, Author(s) Chihiro The ISME Journal, 7(5), 1003-1015 Citation https://doi.org/10.1038/ismej.2012.171 Issue Date 2013-03 Doc URL http://hdl.handle.net/2115/53167 Type article (author version) File Information ISME_Nakao.pdf Instructions for use Hokkaido University Collection of Scholarly and Academic Papers : HUSCAP A novel approach, based on BLSOMs (Batch Learning Self-Organizing Maps), to the microbiome analysis of ticks Ryo Nakao1,a, Takashi Abe2,3,a, Ard M. Nijhof4, Seigo Yamamoto5, Frans Jongejan6,7, Toshimichi Ikemura2, Chihiro Sugimoto1 1Division of Collaboration and Education, Research Center for Zoonosis Control, Hokkaido University, Kita-20, Nishi-10, Kita-ku, Sapporo, Hokkaido 001-0020, Japan 2Nagahama Institute of Bio-Science and Technology, Nagahama, Shiga 526-0829, Japan 3Graduate School of Science & Technology, Niigata University, 8050, Igarashi 2-no-cho, Nishi- ku, Niigata 950-2181, Japan 4Institute for Parasitology and Tropical Veterinary Medicine, Freie Universität Berlin, Königsweg 67, 14163 Berlin, Germany 5Miyazaki Prefectural Institute for Public Health and Environment, 2-3-2 Gakuen Kibanadai Nishi, Miyazaki 889-2155, Japan 6Utrecht Centre for Tick-borne Diseases (UCTD), Department of Infectious Diseases and Immunology, Faculty of Veterinary Medicine, Utrecht University, Yalelaan 1, 3584 CL Utrecht, The Netherlands 7Department of Veterinary Tropical Diseases, Faculty of Veterinary Science, University of Pretoria, Private Bag X04, 0110 Onderstepoort, South Africa aThese authors contributed equally to this work. Keywords: BLSOMs/emerging diseases/metagenomics/microbiomes/symbionts/ticks Running title: Tick microbiomes revealed by BLSOMs Subject category: Microbe-microbe and microbe-host interactions Abstract Ticks transmit a variety of viral, bacterial and protozoal pathogens, which are often zoonotic. -

Babela Massiliensis, a Representative of a Widespread Bacterial
Babela massiliensis, a representative of a widespread bacterial phylum with unusual adaptations to parasitism in amoebae Isabelle Pagnier, Natalya Yutin, Olivier Croce, Kira S Makarova, Yuri I Wolf, Samia Benamar, Didier Raoult, Eugene V. Koonin, Bernard La Scola To cite this version: Isabelle Pagnier, Natalya Yutin, Olivier Croce, Kira S Makarova, Yuri I Wolf, et al.. Babela mas- siliensis, a representative of a widespread bacterial phylum with unusual adaptations to parasitism in amoebae. Biology Direct, BioMed Central, 2015, 10 (13), 10.1186/s13062-015-0043-z. hal-01217089 HAL Id: hal-01217089 https://hal-amu.archives-ouvertes.fr/hal-01217089 Submitted on 19 Oct 2015 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Pagnier et al. Biology Direct (2015) 10:13 DOI 10.1186/s13062-015-0043-z RESEARCH Open Access Babela massiliensis, a representative of a widespread bacterial phylum with unusual adaptations to parasitism in amoebae Isabelle Pagnier1, Natalya Yutin2, Olivier Croce1, Kira S Makarova2, Yuri I Wolf2, Samia Benamar1, Didier Raoult1, Eugene V Koonin2 and Bernard La Scola1* Abstract Background: Only a small fraction of bacteria and archaea that are identifiable by metagenomics can be grown on standard media. -

Characterization of the Interaction Between R. Conorii and Human
Louisiana State University LSU Digital Commons LSU Doctoral Dissertations Graduate School 4-5-2018 Characterization of the Interaction Between R. Conorii and Human Host Vitronectin in Rickettsial Pathogenesis Abigail Inez Fish Louisiana State University and Agricultural and Mechanical College, [email protected] Follow this and additional works at: https://digitalcommons.lsu.edu/gradschool_dissertations Part of the Bacteria Commons, Bacteriology Commons, Biology Commons, Immunology of Infectious Disease Commons, and the Pathogenic Microbiology Commons Recommended Citation Fish, Abigail Inez, "Characterization of the Interaction Between R. Conorii and Human Host Vitronectin in Rickettsial Pathogenesis" (2018). LSU Doctoral Dissertations. 4566. https://digitalcommons.lsu.edu/gradschool_dissertations/4566 This Dissertation is brought to you for free and open access by the Graduate School at LSU Digital Commons. It has been accepted for inclusion in LSU Doctoral Dissertations by an authorized graduate school editor of LSU Digital Commons. For more information, please [email protected]. CHARACTERIZATION OF THE INTERACTION BETWEEN R. CONORII AND HUMAN HOST VITRONECTIN IN RICKETTSIAL PATHOGENESIS A Dissertation Submitted to the Graduate Faculty of the Louisiana State University and Agricultural and Mechanical College in partial fulfillment of the requirements for the degree of Doctor of Philosophy in The Interdepartmental Program in Biomedical and Veterinary Medical Sciences Through the Department of Pathobiological Sciences by Abigail Inez -

(Scrub Typhus). Incubation Period 1 to 3
TYPHUS Causative Agents TYPHUS Rickettsia typhi (murine typhus) and Orientia tsutsugamushi (scrub typhus). Causative Agents IncubationRickettsia typhi Period (murine typhus) and Orientia tsutsugamushi (scrub typhus). 1 to 3 weeks Incubation Period Infectious1 to 3 weeks Period Zoonoses with no human-to-human transmission. Infectious Period TransmissionZoonoses with no human-to-human transmission. Scrub typhus: Bite of grass mites (larval trombiculid mites) MurineTransmission typhus: Bite of rat fleas (also cat and mice fleas) RodentsScrub typhus: are the Bite preferred of grass and mites normal (larval hosts. trombiculid mites) Murine typhus: Bite of rat fleas (also cat and mice fleas) EpidemiologyRodents are the preferred and normal hosts. Distributed throughout the Asia-Pacific rim and is a common cause of pyrexia of unknownEpidemiology origin throughout SE Asia. Occupational contact with rats (e.g. construDistributedction throughout workers inthe makeAsia-Pshiftacific container rim and isfacilities, a common shop cause owners, of pyrexia granary of workers,unknown andorigin garbage throughout collectors) SE orAsia. exposure Occupational to mite habitat contacts in lonwithg grassrats (e.g. hikersconstru andction so ldiers)workers are inrisk make factors.-shift container facilities, shop owners, granary workers, and garbage collectors) or exposure to mite habitats in long grass (e.g. Inhikers Singapore, and soldiers) a total are ofrisk 13 factors. laboratory confirmed cases of murine typhus were r eported in 2008. The majority of cases were foreign workers. In Singapore, a total of 13 laboratory confirmed cases of murine typhus were Clinicalreported Featuresin 2008. The majority of cases were foreign workers. Fever Clinical Headache Features (prominent) MyalgiaFever ConjunctiHeadache val(prominent) suffusion MaculopapularMyalgia rash Conjunctival suffusion Scrub Maculopapular typhus may alsorash have: relative bradycardia, eschar (80%), painful regional adenopathy, hepatosplenomegaly, meningoencephalitis and renal failure. -

BD-CS-057, REV 0 | AUGUST 2017 | Page 1
EXPLIFY RESPIRATORY PATHOGENS BY NEXT GENERATION SEQUENCING Limitations Negative results do not rule out viral, bacterial, or fungal infections. Targeted, PCR-based tests are generally more sensitive and are preferred when specific pathogens are suspected, especially for DNA viruses (Adenovirus, CMV, HHV6, HSV, and VZV), mycobacteria, and fungi. The analytical sensitivity of this test depends on the cellularity of the sample and the concentration of all microbes present. Analytical sensitivity is assessed using Internal Controls that are added to each sample. Sequencing data for Internal Controls is quantified. Samples with Internal Control values below the validated minimum may have reduced analytical sensitivity or contain inhibitors and are reported as ‘Reduced Analytical Sensitivity’. Additional respiratory pathogens to those reported cannot be excluded in samples with ‘Reduced Analytical Sensitivity’. Due to the complexity of next generation sequencing methodologies, there may be a risk of false-positive results. Contamination with organisms from the upper respiratory tract during specimen collection can also occur. The detection of viral, bacterial, and fungal nucleic acid does not imply organisms causing invasive infection. Results from this test need to be interpreted in conjunction with the clinical history, results of other laboratory tests, epidemiologic information, and other available data. Confirmation of positive results by an alternate method may be indicated in select cases. Validated Organisms BACTERIA Achromobacter -

Phenotypic and Genomic Analyses of Burkholderia Stabilis Clinical Contamination, Switzerland Helena M.B
RESEARCH Phenotypic and Genomic Analyses of Burkholderia stabilis Clinical Contamination, Switzerland Helena M.B. Seth-Smith, Carlo Casanova, Rami Sommerstein, Dominik M. Meinel,1 Mohamed M.H. Abdelbary,2 Dominique S. Blanc, Sara Droz, Urs Führer, Reto Lienhard, Claudia Lang, Olivier Dubuis, Matthias Schlegel, Andreas Widmer, Peter M. Keller,3 Jonas Marschall, Adrian Egli A recent hospital outbreak related to premoistened gloves pathogens that generally fall within the B. cepacia com- used to wash patients exposed the difficulties of defining plex (Bcc) (1). Burkholderia bacteria have large, flexible, Burkholderia species in clinical settings. The outbreak strain multi-replicon genomes, a large metabolic repertoire, vari- displayed key B. stabilis phenotypes, including the inabil- ous virulence factors, and inherent resistance to many anti- ity to grow at 42°C; we used whole-genome sequencing to microbial drugs (2,3). confirm the pathogen was B. stabilis. The outbreak strain An outbreak of B. stabilis was identified among hos- genome comprises 3 chromosomes and a plasmid, shar- ing an average nucleotide identity of 98.4% with B. stabilis pitalized patients across several cantons in Switzerland ATCC27515 BAA-67, but with 13% novel coding sequenc- during 2015–2016 (4). The bacterium caused bloodstream es. The genome lacks identifiable virulence factors and has infections, noninvasive infections, and wound contamina- no apparent increase in encoded antimicrobial drug resis- tions. The source of the infection was traced to contaminat- tance, few insertion sequences, and few pseudogenes, ed commercially available, premoistened washing gloves suggesting this outbreak was an opportunistic infection by used for bedridden patients. After hospitals discontinued an environmental strain not adapted to human pathogenic- use of these gloves, the outbreak resolved. -

“Candidatus Deianiraea Vastatrix” with the Ciliate Paramecium Suggests
bioRxiv preprint doi: https://doi.org/10.1101/479196; this version posted November 27, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. The extracellular association of the bacterium “Candidatus Deianiraea vastatrix” with the ciliate Paramecium suggests an alternative scenario for the evolution of Rickettsiales 5 Castelli M.1, Sabaneyeva E.2, Lanzoni O.3, Lebedeva N.4, Floriano A.M.5, Gaiarsa S.5,6, Benken K.7, Modeo L. 3, Bandi C.1, Potekhin A.8, Sassera D.5*, Petroni G.3* 1. Centro Romeo ed Enrica Invernizzi Ricerca Pediatrica, Dipartimento di Bioscienze, Università 10 degli studi di Milano, Milan, Italy 2. Department of Cytology and Histology, Faculty of Biology, Saint Petersburg State University, Saint-Petersburg, Russia 3. Dipartimento di Biologia, Università di Pisa, Pisa, Italy 4 Centre of Core Facilities “Culture Collections of Microorganisms”, Saint Petersburg State 15 University, Saint Petersburg, Russia 5. Dipartimento di Biologia e Biotecnologie, Università degli studi di Pavia, Pavia, Italy 6. UOC Microbiologia e Virologia, Fondazione IRCCS Policlinico San Matteo, Pavia, Italy 7. Core Facility Center for Microscopy and Microanalysis, Saint Petersburg State University, Saint- Petersburg, Russia 20 8. Department of Microbiology, Faculty of Biology, Saint Petersburg State University, Saint- Petersburg, Russia * Corresponding authors, contacts: [email protected] ; [email protected] 1 bioRxiv preprint doi: https://doi.org/10.1101/479196; this version posted November 27, 2018. -

Fleas and Flea-Borne Diseases
International Journal of Infectious Diseases 14 (2010) e667–e676 Contents lists available at ScienceDirect International Journal of Infectious Diseases journal homepage: www.elsevier.com/locate/ijid Review Fleas and flea-borne diseases Idir Bitam a, Katharina Dittmar b, Philippe Parola a, Michael F. Whiting c, Didier Raoult a,* a Unite´ de Recherche en Maladies Infectieuses Tropicales Emergentes, CNRS-IRD UMR 6236, Faculte´ de Me´decine, Universite´ de la Me´diterrane´e, 27 Bd Jean Moulin, 13385 Marseille Cedex 5, France b Department of Biological Sciences, SUNY at Buffalo, Buffalo, NY, USA c Department of Biology, Brigham Young University, Provo, Utah, USA ARTICLE INFO SUMMARY Article history: Flea-borne infections are emerging or re-emerging throughout the world, and their incidence is on the Received 3 February 2009 rise. Furthermore, their distribution and that of their vectors is shifting and expanding. This publication Received in revised form 2 June 2009 reviews general flea biology and the distribution of the flea-borne diseases of public health importance Accepted 4 November 2009 throughout the world, their principal flea vectors, and the extent of their public health burden. Such an Corresponding Editor: William Cameron, overall review is necessary to understand the importance of this group of infections and the resources Ottawa, Canada that must be allocated to their control by public health authorities to ensure their timely diagnosis and treatment. Keywords: ß 2010 International Society for Infectious Diseases. Published by Elsevier Ltd. All rights reserved. Flea Siphonaptera Plague Yersinia pestis Rickettsia Bartonella Introduction to 16 families and 238 genera have been described, but only a minority is synanthropic, that is they live in close association with The past decades have seen a dramatic change in the geographic humans (Table 1).4,5 and host ranges of many vector-borne pathogens, and their diseases. -
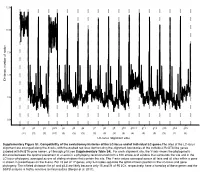
LC-Locus Alignment Sites Distance, Number of Nodes Supplementary
12.5 10.0 7.5 5.0 Distance, number of nodes 2.5 0.0 g1 g2 g3 g3.5 g4 g5 g6 g7 g8 g9 g10 g10.1 g11 g12 g13 g14 g15 (11) (3) (3) (10) (3) (3) (3) (3) (3) (3) (3) (4) (3) (3) (3) (2) (3) LC-locus alignment sites Supplementary Figure S1. Compatibility of the evolutionary histories of the LC-locus and of individual LC genes.The sites of the LC-locus alignment are arranged along the X-axis, with the dashed red lines demarcating the alignment boundaries of the individual RcGTA-like genes (labeled with RcGTA gene names, g1 through g15; see Supplementary Table S4). For each alignment site, the Y-axis shows the phylogenetic distance between the optimal placement of a taxon in a phylogeny reconstructed from a 100 amino-acid window that surrounds the site and in the LC-locus phylogeny, averaged across all sliding windows that contain the site. The Y-axis values averaged across all taxa and all sites within a gene is shown in parentheses on the X-axis. For 15 out of 17 genes, only 2-4 nodes separate the optimal taxon position in the LC-locus and gene phylogeny. The inflated distances for g1 and g3.5 are likely because only 15 and 21 of 95 LCs, respectively, have a homolog of these genes and the SSPB analysis is highly sensitive to missing data (Berger et al. 2011). a. Bacteria Unassigned Thermotogae Tenericutes Synergistetes Spirochaetes Proteobacteria ylum Planctomycetes h p Firmicutes Deferribacteres Cyanobacteria Chloroflexi Bacteroidetes Actinobacteria Acidobacteria 1(11,750) 2(1,750) 3(2,538) 4(168) 5(51) 6(54) 7(43) 8(32) 9(26) 10(40) 11(33) 12(198) 13(173) 14(101) 15(98) 16(43) 17(114) Number of rcc01682−rcc01698 homologs in a cluster b. -

Genome Project Reveals a Putative Rickettsial Endosymbiont
GBE Bacterial DNA Sifted from the Trichoplax adhaerens (Animalia: Placozoa) Genome Project Reveals a Putative Rickettsial Endosymbiont Timothy Driscoll1,y, Joseph J. Gillespie1,2,*,y, Eric K. Nordberg1,AbduF.Azad2, and Bruno W. Sobral1,3 1Virginia Bioinformatics Institute at Virginia Polytechnic Institute and State University 2Department of Microbiology and Immunology, University of Maryland School of Medicine 3Present address: Nestle´ Institute of Health Sciences SA, Campus EPFL, Quartier de L’innovation, Lausanne, Switzerland *Corresponding author: E-mail: [email protected]. yThese authors contributed equally to this work. Accepted: March 1, 2013 Abstract Eukaryotic genome sequencing projects often yield bacterial DNA sequences, data typically considered as microbial contamination. However, these sequences may also indicate either symbiont genes or lateral gene transfer (LGT) to host genomes. These bacterial sequences can provide clues about eukaryote–microbe interactions. Here, we used the genome of the primitive animal Trichoplax adhaerens (Metazoa: Placozoa), which is known to harbor an uncharacterized Gram-negative endosymbiont, to search for the presence of bacterial DNA sequences. Bioinformatic and phylogenomic analyses of extracted data from the genome assembly (181 bacterial coding sequences [CDS]) and trace read archive (16S rDNA) revealed a dominant proteobacterial profile strongly skewed to Rickettsiales (Alphaproteobacteria) genomes. By way of phylogenetic analysis of 16S rDNA and 113 proteins conserved across proteobacterial genomes, as well as identification of 27 rickettsial signature genes, we propose a Rickettsiales endosymbiont of T. adhaerens (RETA). The majority (93%) of the identified bacterial CDS belongs to small scaffolds containing prokaryotic-like genes; however, 12 CDS were identified on large scaffolds comprised of eukaryotic-like genes, suggesting that T. -

Rickettsialpox-A Newly Recognized Rickettsial Disease V
Public Health Reports Vol. 62 * MAY 30, 1947 * No. 22 Printed With the Approval of the Bureau of the Budget as Required by Rule 42 of the Joint-Committee on Printing RICKETTSIALPOX-A NEWLY RECOGNIZED RICKETTSIAL DISEASE V. RECOVERY OF RICKETTSIA AKARI FROM A HOUSE MOUSE (MUS MUSCULUS)1 By ROBERT J. HUEBNER, Senior Assistant Surgeon, WILLIAm L. JELLISON, Parasitologist, CHARLES ARMSTRONG, Medical Director, United States Public Health Service Ricketttia akari, the causative agent of rickettsialpox, was isolated from the blood of persons ill with this disease (1) and from rodent mites Allodermanyssus sanguineus Hirst inhabiting the domicile of ill per- sons (2). This paper describes the isolation of R. akari from a house mouse (Mus musculus) trapped on the same premises-a housing development in the citr of New York where more than 100 cases of rickettsialpox have occurred (3), (4), (5), (6). Approximately 60 house mice were trapped in the basements of this housing development where rodent harborage existed in store rooms and in incinerator ashpits. Engorged mites were occasionally found attached to the mice, the usual site of attachment being the rump. Mites were frequently found inside the box traps after the captured mice were removed. Early attempts to isolate the etiological agent of rickettisalpox from these mice were complicated by the presence of choriomeningitis among them. Twelve successive suspensions of mouse tissue, repre- senting 16 house mice, inoculated intracerebrally into laboratory mice (Swiss strain) and intraperitoneally into guinea pigs resulted in the production of a highly lethal disease in both species which was identified immunologically as choriomeningitis. -

Ohio Department of Health, Bureau of Infectious Diseases Disease Name Class A, Requires Immediate Phone Call to Local Health
Ohio Department of Health, Bureau of Infectious Diseases Reporting specifics for select diseases reportable by ELR Class A, requires immediate phone Susceptibilities specimen type Reportable test name (can change if Disease Name other specifics+ call to local health required* specifics~ state/federal case definition or department reporting requirements change) Culture independent diagnostic tests' (CIDT), like BioFire panel or BD MAX, E. histolytica Stain specimen = stool, bile results should be sent as E. histolytica DNA fluid, duodenal fluid, 260373001^DETECTED^SCT with E. histolytica Antigen Amebiasis (Entamoeba histolytica) No No tissue large intestine, disease/organism-specific DNA LOINC E. histolytica Antibody tissue small intestine codes OR a generic CIDT-LOINC code E. histolytica IgM with organism-specific DNA SNOMED E. histolytica IgG codes E. histolytica Total Antibody Ova and Parasite Anthrax Antibody Anthrax Antigen Anthrax EITB Acute Anthrax EITB Convalescent Anthrax Yes No Culture ELISA PCR Stain/microscopy Stain/spore ID Eastern Equine Encephalitis virus Antibody Eastern Equine Encephalitis virus IgG Antibody Eastern Equine Encephalitis virus IgM Arboviral neuroinvasive and non- Eastern Equine Encephalitis virus RNA neuroinvasive disease: Eastern equine California serogroup virus Antibody encephalitis virus disease; LaCrosse Equivocal results are accepted for all California serogroup virus IgG Antibody virus disease (other California arborviral diseases; California serogroup virus IgM Antibody specimen = blood, serum, serogroup