Building a New Biology
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Genomic Misconception
1 Genomic Misconception: A fresh look at the biosafety of transgenic and conventional crops. a plea for a process agnostic regulation Klaus Ammann, University of Bern, [email protected] , Neuchâtel, Switzerland December 9, 2012 names. A shorter version has been published in New Biotechnology Ammann, K. (2014), Genomic Misconception: a fresh look at the biosafety of transgenic and conventional crops. A plea for a process agnostic regulation, New Biotechnology, 31, 1, pp. 1-17, http://www.ask-force.org/web/NewBiotech/Ammann-Genomic-Misconception-printed-2014.pdf Abstract: .......................................................................................................................................... 1 1. The scientific basis of the process agnostic regulation ................................................................... 2 2. How the Genomic Misconception was evolving ............................................................................. 4 2.1. How the ‘Genomic Misconception’ was erroneously maintained in the European Regulation and the Cartagena Protocol on Biosafety ....................................................................................... 6 a) Europe, the development of the transatlantic divide with the United States ........................ 6 b) Legislative History of the Cartagena Protocol and its Genomic Misinterpretation of Transgenesis ................................................................................................................................ 8 3. Conclusions ................................................................................................................................... -

Spread of a SARS-Cov-2 Variant Through Europe in the Summer of 2020
Article Spread of a SARS-CoV-2 variant through Europe in the summer of 2020 https://doi.org/10.1038/s41586-021-03677-y Emma B. Hodcroft1,2,3 ✉, Moira Zuber1, Sarah Nadeau2,4, Timothy G. Vaughan2,4, Katharine H. D. Crawford5,6,7, Christian L. Althaus3, Martina L. Reichmuth3, John E. Bowen8, Received: 25 November 2020 Alexandra C. Walls8, Davide Corti9, Jesse D. Bloom5,6,10, David Veesler8, David Mateo11, Accepted: 28 May 2021 Alberto Hernando11, Iñaki Comas12,13, Fernando González-Candelas13,14, SeqCOVID-SPAIN consortium*, Tanja Stadler2,4,92 & Richard A. Neher1,2,92 ✉ Published online: 7 June 2021 Check for updates Following its emergence in late 2019, the spread of SARS-CoV-21,2 has been tracked by phylogenetic analysis of viral genome sequences in unprecedented detail3–5. Although the virus spread globally in early 2020 before borders closed, intercontinental travel has since been greatly reduced. However, travel within Europe resumed in the summer of 2020. Here we report on a SARS-CoV-2 variant, 20E (EU1), that was identifed in Spain in early summer 2020 and subsequently spread across Europe. We fnd no evidence that this variant has increased transmissibility, but instead demonstrate how rising incidence in Spain, resumption of travel, and lack of efective screening and containment may explain the variant’s success. Despite travel restrictions, we estimate that 20E (EU1) was introduced hundreds of times to European countries by summertime travellers, which is likely to have undermined local eforts to minimize infection with SARS-CoV-2. Our results illustrate how a variant can rapidly become dominant even in the absence of a substantial transmission advantage in favourable epidemiological settings. -

Steven Henikoff Position
CURRICULUM VITAE: Steven Henikoff Position: Investigator, Howard Hughes Medical Institute Member, Basic Sciences Division Address: Fred Hutchinson Cancer Research Center 1100 Fairview Ave N. Seattle, Washington 98109-1024 Phone (206) 667-4515; FAX (206) 667-5889 E-mail: [email protected] http://blocks.fhcrc.org/~steveh/ Education 1964-68 University of Chicago, Chicago, Illinois. BS in Chemistry. Research on optical properties of biopolymers, Dr. G. Holzwarth, advisor. 1971-77 Harvard University, Cambridge, Massachusetts. PhD in Biochemistry and Molecular Biology. Dr. M. Meselson, advisor. Thesis: RNA from heat induced puff sites in Drosophila. 1977-80 University of Washington, Seattle, Washington. Postdoctoral fellow in Zoology. Research on position-effect variegation in Drosophila, Dr. C. Laird, advisor, Leukemia Society of America fellow. Professional Experience 1981-85 Fred Hutchinson Cancer Research Center, Seattle, Washington. Assistant Member in Basic Sciences. 1981- University of Washington, Seattle. Affiliate Assistant, Associate and Full Professor of Genetics/Genome Sciences. 1985-88 Fred Hutchinson Cancer Research Center, Seattle, Washington. Associate Member in Basic Sciences. 1988- Fred Hutchinson Cancer Research Center, Seattle, Washington. Member in Basic Sciences. 1990- Investigator, Howard Hughes Medical Institute. Current Research Nucleosome dynamics Transcriptional regulation Centromeric chromatin and centromere evolution Epigenomic technologies Honors (since 2000) 2001 Keynote, 13th International Arabidopsis Conference, -

Developmental Biology Using Purified Genes
LASKER~KOSHLAND SPECIAL ACHIEVEMENT ESSAY IN MEDICAL SCIENCE AWARD Developmental biology using purified genes Donald D Brown Some history Control Anucleolate Magnesium From the NIH I went to the Pasteur Institute mutant deficient After three years of college I entered the in Paris to study bacterial gene regulation in University of Chicago Medical School in the 1959, the year after the Lac repressor had been fall of 1952 and discovered biochemistry and discovered. Before leaving Bethesda, by the research. Lloyd Kozloff, a member of the greatest luck I learned about a small research bacteriophage group in the Department of institution in Baltimore that was associated Biochemistry, guided my research. While in at that time with the Johns Hopkins Medical medical school I began searching for a future School called the Department of Embryology of research subject, thinking it should be an the Carnegie Institution of Washington. I con- important medically related problem but unex- tacted Jim Ebert, the director, and arranged an plored by what were then the modern methods advanced postdoctoral fellowship after my year of biochemistry. in Paris. It is hard to imagine two more diverse The field of embryology, newly named research institutions. ‘developmental biology’, caught my attention. The Pasteur Institute was at the forefront of Reproductive biology was barely discussed, biology, involved in the founding of molecular and descriptive embryology was taught in two biology. Every day at lunch Jacques Monod, lectures as a part of gross anatomy. In 1953, I François Jacob and André Lwoff presided attended a biochemistry journal club discus- over an exciting discussion usually augmented Figure 1 Comparison of control (left), anucleolate sion of the Watson-Crick Nature paper describ- by a prominent visitor. -

Enhancers, Enhancers – from Their Discovery to Today’S Universe of Transcription Enhancers
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by RERO DOC Digital Library Biol. Chem. 2015; 396(4): 311–327 Review Walter Schaffner* Enhancers, enhancers – from their discovery to today’s universe of transcription enhancers Abstract: Transcriptional enhancers are short (200–1500 one had ever postulated their existence, simply because base pairs) DNA segments that are able to dramatically there seemed to be no need for them. Now that introns boost transcription from the promoter of a target gene. and enhancers are part of the scientific world, one cannot Originally discovered in simian virus 40 (SV40), a small imagine how higher forms of life could ever have evolved DNA virus, transcription enhancers were soon also found without the multitude of tailored proteins that can be in immunoglobulin genes and other cellular genes as produced by alternative splicing, or without the sophisti- key determinants of cell-type-specific gene expression. cated patterns of remote transcription control by enhanc- Enhancers can exert their effect over long distances of ers. Indeed, the complexity of an organism is primarily thousands, even hundreds of thousands of base pairs, determined by the variety of gene regulation mechanisms, either from upstream, downstream, or from within a tran- rather than by the number of genes. scription unit. The number of enhancers in eukaryotic genomes correlates with the complexity of the organism; a typical mammalian gene is likely controlled by several enhancers to fine-tune its expression at different devel- The holy grail opmental stages, in different cell types and in response In the fall of 1978, I returned to Zurich University from to different signaling cues. -

Steven Henikoff Position
CURRICULUM VITAE: Steven Henikoff Position: Investigator, Howard Hughes Medical Institute Member, Basic Sciences Division Address: Fred Hutchinson Cancer Research Center 1100 Fairview Ave N. Seattle, Washington 98109-1024 Phone (206) 667-4515; FAX (206) 667-5889 E-mail: [email protected] http://research.fhcrc.org/henikoff/en.html Education 1964-68 UniversitY of Chicago, Chicago, Illinois. BS in ChemistrY. Research on optical properties of biopolymers, Dr. G. Holzwarth, advisor. 1971-77 Harvard UniversitY, Cambridge, Massachusetts. PhD in BiochemistrY and Molecular BiologY. Dr. M. Meselson, advisor. Thesis: RNA from heat induced puff sites in Drosophila. 1977-80 UniversitY of Washington, Seattle, Washington. Postdoctoral fellow in Zoology. Research on position-effect variegation in Drosophila, Dr. C. Laird, advisor, Leukemia SocietY of America fellow. Professional Experience 1981-85 Fred Hutchinson Cancer Research Center, Seattle, Washington. Assistant Member in Basic Sciences. 1981- UniversitY of Washington, Seattle. Affiliate Assistant, Associate and Full Professor of Genetics/Genome Sciences. 1985-88 Fred Hutchinson Cancer Research Center, Seattle, Washington. Associate Member in Basic Sciences. 1988- Fred Hutchinson Cancer Research Center, Seattle, Washington. Member in Basic Sciences. 1990- Investigator, Howard Hughes Medical Institute. Current Research Nucleosome dYnamics Transcriptional regulation Centromeric chromatin and centromere evolution Epigenomic technologies Ongoing Funding Howard Hughes Medical Institute Investigator 04/1/1990 -
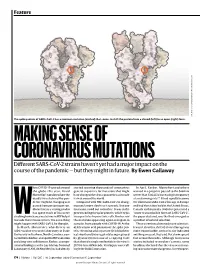
MAKING SENSE of CORONAVIRUS MUTATIONS Different SARS-Cov-2 Strains Haven’T Yet Had a Major Impact on the Course of the Pandemic — but They Might in Future
Feature SOURCE: STRUCTURAL DATA FROM K. SHEN & J. LUBAN K. SHEN FROM DATA STRUCTURAL SOURCE: The spike protein of SARS-CoV-2 has a common mutation (circled) that seems to shift the protein from a closed (left) to an open (right) form. MAKING SENSE OF CORONAVIRUS MUTATIONS Different SARS-CoV-2 strains haven’t yet had a major impact on the course of the pandemic — but they might in future. By Ewen Callaway hen COVID-19 spread around started scouring thousands of coronavirus In April, Korber, Montefiori and others the globe this year, David genetic sequences for mutations that might warned in a preprint posted to the bioRxiv Montefiori wondered how the have changed the virus’s properties as it made server that “D614G is increasing in frequency deadly virus behind the pan- its way around the world. at an alarming rate”1. It had rapidly become demic might be changing as it Compared with HIV, SARS-CoV-2 is chang- the dominant SARS-CoV-2 lineage in Europe passed from person to person. ing much more slowly as it spreads. But one and had then taken hold in the United States, Montefiori is a virologist who mutation stood out to Korber. It was in the Canada and Australia. D614G represented a has spent much of his career gene encoding the spike protein, which helps “more transmissible form of SARS-CoV-2”, Wstudying how chance mutations in HIV help it virus particles to penetrate cells. Korber saw the paper declared, one that had emerged as to evade the immune system. -

Annual Report 2000
ST. KILDA SOAY SHEEP PROJECT: ANNUAL REPORT 2000 J.G. Pilkington1+3, S.D. Albon2, W. Amos3, D.W. Coltman1+8, T.H. Clutton- Brock3, T.N. Coulson3, M.J. Crawley4, M.C. Forchhammer3, I. Gordon5, B.T. Grenfell3, M. Hutchings5, A.W. Illius1, O. Jones4+5, L. Kruuk1, J. Lindström3+7, J.M. Pemberton1, B. Preston6, I.R. Stevenson6, K. Wilson6. 1Institute of Cell, Animal and Population Biology, University of Edinburgh. 2Centre for Ecology and Hydrology, Banchory 3Department of Zoology, University of Cambridge. 4Department of Biology, Imperial College, University of London. 5Macaulay Land Use Research Institute, Aberdeen. 6Institute of Biological Sciences, University of Stirling. New addresses – 7Institute of Biomedical and Life Sciences, University of Glasgow. 8Department of Animal and Plant Sciences, University of Sheffield. POPULATION OVERVIEW..................................................................................................................................... 2 REPORTS ON COMPONENT STUDIES .................................................................................................................... 4 Reproductive energetics of rams, and the maintenance of horn diversity .................................................... 4 Ram behaviour with respect to "favoured" insemination times .................................................................... 5 Sperm depletion in Soay rams....................................................................................................................... 7 Vegetation .................................................................................................................................................... -

Estimation of Genetic Variance in Fitness, and Inference of Adaptation, When Fitness Follows a Log-Normal Distribution Timothée Bonnet , Michael B
Journal of Heredity, 2019, 1–13 doi:10.1093/jhered/esz018 THE 2017 AGA KEY DISTINGUISHED LECTURE Advance Access publication June 26, 2019 Downloaded from https://academic.oup.com/jhered/advance-article-abstract/doi/10.1093/jhered/esz018/5523732 by Da-Collect Chifley Library ANUC user on 01 July 2019 THE 2017 AGA KEY DISTINGUISHED LECTURE Estimation of Genetic Variance in Fitness, and Inference of Adaptation, When Fitness Follows a Log-Normal Distribution Timothée Bonnet , Michael B. Morrissey, and Loeske E. B. Kruuk From the Division of Ecology and Evolution, Research School of Biology, The Australian National University, 46 Sullivans Creek Road, Canberra, ACT 2600, Australia (Bonnet and Kruuk); and School of Biology, University of St Andrews, St Andrews Fife KY16 9TH, UK (Morrissey). Address correspondence to Timothée Bonnet at the address above, or e-mail: [email protected]. Received November 14, 2018; First decision January 23, 2019; Accepted April 7, 2019. Corresponding Editor: Anne Bronikowski Loeske Kruuk is a professor in the Research School of Biology at the Australian National University. She did a PhD in population genetics at the University of Edinburgh, followed by a postdoc at the University of Cambridge, and then held a Royal Society University Research Fellowship at the University of Edinburgh and an Australian Research Council Future Fellowship at the Australian National University. Her research focuses on evolutionary ecology and quantitative genetics of natural populations, with the aim of understanding how both evolutionary processes and environmental conditions shape diversity in natural populations. She has worked on a range of species, largely but not entirely using long-term studies of wild vertebrate populations with individual-level field data. -

DNA Methylation Patterns and Cancer
restriction/modification system, which brought Werner Arber, Daniel Nathans and Hamilton Smith the 1978 Nobel Prize in Physiology or Medicine, and made restriction enzymes the primary tools of Charles Rodolphe Brupbacher Foundation molecular biology. Four decades have passed since then, but the role of 5-methylcytosine in eukaryotic DNA metabolism is still shrouded in mystery. We know that the sperm methylation pattern is largely The erased after fertilization and that methylation is gradually reintroduced Charles Rodolphe Brupbacher Prize during embryogenesis and differentiation, but the processes that for Cancer Research 2017 regulate the cell type- and tissue-specific methylation patterns remain is awarded to to be elucidated. We have also learned that DNA can be not only methylated, but also demethylated, and that aberrant methylation can lead to disease - including cancer. Again, how these processes are regulated remains to be discovered. However, we have learnt a great Sir Adrian Peter Bird, deal about 5-methylcytosine metabolism during the past three decades and much of our knowledge came from the laboratory of Adrian Bird. PhD Adrian spent his doctoral and postdoctoral time in Max Birnstiel’s for his contributions to our understanding laboratory, first in Edinburgh and then in Zurich, studying the amplification of ribosomal DNA in Xenopus laevis. In this organism, of the role of DNA methylation in genomic rDNA in somatic tissues is highly methylated, while the development and disease extrachromosomal amplicons are unmethylated. When he returned to Edinburgh to establish his own group, Adrian set out to study The President The President of the Foundation of the Scientific Advisory Board the methylation pattern of these loci using the newly-available methylation-sensitive restriction enzymes. -

Evolutionary Dynamics and Fitness in Wild Populations
2018 © II Joint Congress on Evolutionary Biology 2018. All rights reserved - Any reproductionEvolutionary even in part is prohibited. dynamics and fitness in wild populations 2018 © II Joint Congress on Evolutionary Biology 2018. All rights reserved - Any reproduction even in part is prohibited. 2018 © 48th European Contact Lens Society Of Ophthalmologists. All rights reserved - Any reproduction even in part is prohibited] . Loeske Kruuk 2018 © II Joint Congress on Evolutionary Biology 2018. All rights reserved - Any reproduction even in part is prohibited] . 2018 © II Joint Congress on Evolutionary Biology 2018. All rights reserved - Any reproduction even in part is prohibited] . 2018 © II Joint Congress on Evolutionary Biology 2018. All rights reserved - Any reproduction even in part is prohibited] . 2018 © II Joint Congress on Evolutionary Biology 2018. All rights reserved - Any reproduction even in part is prohibited] . 2018 © II Joint Congress on Evolutionary Biology 2018. All rights reserved - Any reproduction even in part is prohibited] . 2018 © II Joint Congress on Evolutionary Biology 2018. All rights reserved - Any reproduction even in part is prohibited] . Charles Darwin, Origin of Species 1859 “ No one supposes that all the individuals of the same species are cast in the same actual mould. These individual differences are of the highest importance for us, 2018 © II Joint Congress on Evolutionary Biology 2018. All rights reserved - Any reproduction even in part is prohibited. for they are often inherited …and they thus afford materials for natural selection to act on. ” evolutionary change: genetics and selection 2018 © II Joint Congress on Evolutionary Biology 2018. All rights reserved - Any reproduction even in part is prohibited. -

Evolution and Origins of SARS- Cov-2 and Related Coronaviruses Introduction to Evolution and Scientific Inquiry Spring 2020, Dr
Evolution and origins of SARS- CoV-2 and related coronaviruses Introduction to Evolution and Scientific Inquiry Spring 2020, Dr. Spielman Zoonosis Image from April 2019 • Virus that normally infects one animal species infects a NEW/DIFFERENT species • Sometimes from a new mutation • Sometimes from an existing mutation • Sometimes no mutation at all – it just can infect multiple species Viruses bind host receptors or other cell-surface proteins https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.3000217 ACE2: Angiotension-converting enzyme 2 • ACE constricts blood flow • ACE inhibitors relax veins to improve blood flow. BRIEF COMMUNICATION Used to treat… https://doi.org/10.1038/s41591-020-0868-6 • Heart disease • Angiotension SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate • Diabetes immune genes • Migraines Waradon Sungnak! !1 , Ni Huang1, Christophe Bécavin! !2, Marijn Berg3,4, Rachel Queen5, Monika Litvinukova1,6, Carlos Talavera-López1, Henrike Maatz6, Daniel Reichart7, • Chronic kidney diseases Fotios Sampaziotis! !8,9,10, Kaylee B. Worlock11, Masahiro Yoshida! !11, Josephine L. Barnes11 and HCA Lung Biological Network* We investigated SARS-CoV-2 potential tropism by survey- associated with SARS-CoV-2 pathogenesis at cellular resolution, inghttps:// expressionwww.nature.com of viral entry-associated/articles/s41591 genes in single-cell using-020 single-cell-0868 RNA- 6sequencing (scRNA-seq) datasets from healthy RNA-sequencing data from multiple tissues from healthy donors generated by the Human Cell Atlas consortium and other human donors. We co-detected these transcripts in specific resources to inform and prioritize the use of precious, limited clinical respiratory, corneal and intestinal epithelial cells, potentially material that is becoming available from COVID-19 patients.