Molecular Epidemiology and Evolutionary Trajectory of Emerging Echovirus 30, Europe Kimberley S.M
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Spread of a SARS-Cov-2 Variant Through Europe in the Summer of 2020
Article Spread of a SARS-CoV-2 variant through Europe in the summer of 2020 https://doi.org/10.1038/s41586-021-03677-y Emma B. Hodcroft1,2,3 ✉, Moira Zuber1, Sarah Nadeau2,4, Timothy G. Vaughan2,4, Katharine H. D. Crawford5,6,7, Christian L. Althaus3, Martina L. Reichmuth3, John E. Bowen8, Received: 25 November 2020 Alexandra C. Walls8, Davide Corti9, Jesse D. Bloom5,6,10, David Veesler8, David Mateo11, Accepted: 28 May 2021 Alberto Hernando11, Iñaki Comas12,13, Fernando González-Candelas13,14, SeqCOVID-SPAIN consortium*, Tanja Stadler2,4,92 & Richard A. Neher1,2,92 ✉ Published online: 7 June 2021 Check for updates Following its emergence in late 2019, the spread of SARS-CoV-21,2 has been tracked by phylogenetic analysis of viral genome sequences in unprecedented detail3–5. Although the virus spread globally in early 2020 before borders closed, intercontinental travel has since been greatly reduced. However, travel within Europe resumed in the summer of 2020. Here we report on a SARS-CoV-2 variant, 20E (EU1), that was identifed in Spain in early summer 2020 and subsequently spread across Europe. We fnd no evidence that this variant has increased transmissibility, but instead demonstrate how rising incidence in Spain, resumption of travel, and lack of efective screening and containment may explain the variant’s success. Despite travel restrictions, we estimate that 20E (EU1) was introduced hundreds of times to European countries by summertime travellers, which is likely to have undermined local eforts to minimize infection with SARS-CoV-2. Our results illustrate how a variant can rapidly become dominant even in the absence of a substantial transmission advantage in favourable epidemiological settings. -
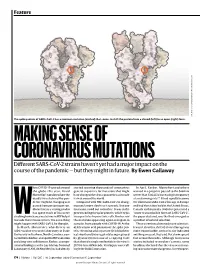
MAKING SENSE of CORONAVIRUS MUTATIONS Different SARS-Cov-2 Strains Haven’T Yet Had a Major Impact on the Course of the Pandemic — but They Might in Future
Feature SOURCE: STRUCTURAL DATA FROM K. SHEN & J. LUBAN K. SHEN FROM DATA STRUCTURAL SOURCE: The spike protein of SARS-CoV-2 has a common mutation (circled) that seems to shift the protein from a closed (left) to an open (right) form. MAKING SENSE OF CORONAVIRUS MUTATIONS Different SARS-CoV-2 strains haven’t yet had a major impact on the course of the pandemic — but they might in future. By Ewen Callaway hen COVID-19 spread around started scouring thousands of coronavirus In April, Korber, Montefiori and others the globe this year, David genetic sequences for mutations that might warned in a preprint posted to the bioRxiv Montefiori wondered how the have changed the virus’s properties as it made server that “D614G is increasing in frequency deadly virus behind the pan- its way around the world. at an alarming rate”1. It had rapidly become demic might be changing as it Compared with HIV, SARS-CoV-2 is chang- the dominant SARS-CoV-2 lineage in Europe passed from person to person. ing much more slowly as it spreads. But one and had then taken hold in the United States, Montefiori is a virologist who mutation stood out to Korber. It was in the Canada and Australia. D614G represented a has spent much of his career gene encoding the spike protein, which helps “more transmissible form of SARS-CoV-2”, Wstudying how chance mutations in HIV help it virus particles to penetrate cells. Korber saw the paper declared, one that had emerged as to evade the immune system. -

Evolution and Origins of SARS- Cov-2 and Related Coronaviruses Introduction to Evolution and Scientific Inquiry Spring 2020, Dr
Evolution and origins of SARS- CoV-2 and related coronaviruses Introduction to Evolution and Scientific Inquiry Spring 2020, Dr. Spielman Zoonosis Image from April 2019 • Virus that normally infects one animal species infects a NEW/DIFFERENT species • Sometimes from a new mutation • Sometimes from an existing mutation • Sometimes no mutation at all – it just can infect multiple species Viruses bind host receptors or other cell-surface proteins https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.3000217 ACE2: Angiotension-converting enzyme 2 • ACE constricts blood flow • ACE inhibitors relax veins to improve blood flow. BRIEF COMMUNICATION Used to treat… https://doi.org/10.1038/s41591-020-0868-6 • Heart disease • Angiotension SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate • Diabetes immune genes • Migraines Waradon Sungnak! !1 , Ni Huang1, Christophe Bécavin! !2, Marijn Berg3,4, Rachel Queen5, Monika Litvinukova1,6, Carlos Talavera-López1, Henrike Maatz6, Daniel Reichart7, • Chronic kidney diseases Fotios Sampaziotis! !8,9,10, Kaylee B. Worlock11, Masahiro Yoshida! !11, Josephine L. Barnes11 and HCA Lung Biological Network* We investigated SARS-CoV-2 potential tropism by survey- associated with SARS-CoV-2 pathogenesis at cellular resolution, inghttps:// expressionwww.nature.com of viral entry-associated/articles/s41591 genes in single-cell using-020 single-cell-0868 RNA- 6sequencing (scRNA-seq) datasets from healthy RNA-sequencing data from multiple tissues from healthy donors generated by the Human Cell Atlas consortium and other human donors. We co-detected these transcripts in specific resources to inform and prioritize the use of precious, limited clinical respiratory, corneal and intestinal epithelial cells, potentially material that is becoming available from COVID-19 patients. -
Midair Scare Brings Probe of 777S Turer Is Increasingly Hosting Crazereaches Beyond Shares Drive-Through Job Fairsand Rais- Favoredonsocial Media
P2JW053000-6-A00100-27FFFF5178F ADVERTISEMENT Meetour new, easy-to-use trading platform: thinkorswim® Web. LearnmoreonpageR10. ****** MONDAY,FEBRUARY 22, 2021 ~VOL. CCLXXVII NO.42 WSJ.com HHHH $4.00 Last week: DJIA 31494.32 À 35.92 0.1% NASDAQ 13874.46 g 1.6% STOXX 600 414.88 À 0.2% 10-YR. TREASURY g 1 11/32 , yield 1.344% OIL $59.24 g $0.23 EURO $1.2120 YEN 105.43 Texas Thaws Out and Repairs Begin on Damage From the Cold Housing, What’s News Online Shopping Business&Finance Propel trength in housing and e- Scommerce during the pan- demic has helped propel hiring JobGains in blue-collar jobs,with em- ployment in some categories exceeding precrisis levels. A1 Blue-collar workers The jump this month in see boom, but some U.S. government-bond yields is sending tremors companies can’t find through stocks and forcing enough qualified help investorstomoreseriously confront the implications BY SARAH CHANEY CAMBON of rising interest rates. A1 Agroup of activist inves- GES TheU.S.’sblue-collar work- tors has abig stakeinKohl’s IMA forcehas begun to benefit from and is attempting to take astrengthening job market. control of the department- GETTY An Orlando,Fla.-area home store chain’s board. B1 AN/ builderisseeking to add four construction workerstoasix- M&T Bank is nearing a SULLIV person team in the midst of deal to acquire People’s TIN soaring housing demand during United Financial for more JUS SHELTER: Plumber Randy Calazans repaired a burst pipe in a home in HoustoN on Sunday, as temperatures climbed and the pandemic.InAtlanta, a than $7 billion, in the lat- Texas started to recover from severe weather that overwhelmed its power grid, leaving homes without heat for days. -

This Thesis Has Been Submitted in Fulfilment of the Requirements for a Postgraduate Degree (E.G
This thesis has been submitted in fulfilment of the requirements for a postgraduate degree (e.g. PhD, MPhil, DClinPsychol) at the University of Edinburgh. Please note the following terms and conditions of use: • This work is protected by copyright and other intellectual property rights, which are retained by the thesis author, unless otherwise stated. • A copy can be downloaded for personal non-commercial research or study, without prior permission or charge. • This thesis cannot be reproduced or quoted extensively from without first obtaining permission in writing from the author. • The content must not be changed in any way or sold commercially in any format or medium without the formal permission of the author. • When referring to this work, full bibliographic details including the author, title, awarding institution and date of the thesis must be given. Transmission Dynamics of Avian Influenza A Virus Lu Lu Submitted for the degree of Doctor of Philosophy University of Edinburgh 2015 Declaration This thesis has been composed by my own. The work is my own work except where otherwise indicated in the text. The work has not been submitted for any other degree or professional qualification except as specified. Lu Lu 2014.12.18 Acknowledgements I appreciate my funding from the China Scholarship Council/University of Edinburgh Joint Scholarship. This work could not have been completed without it. I would like to thank many people who provided great help to not only my PhD study but the life in UK during the past three years. They have made it possible for me to complete this thesis. -

SARS-Cov-2 Variants of Interest and Concern Naming Scheme Conducive for Global Discourse
comment SARS-CoV-2 Variants of Interest and Concern naming scheme conducive for global discourse A group convened and led by the Virus Evolution Working Group of the World Health Organization reports on its deliberations and announces a naming scheme that will enable clear communication about SARS-CoV-2 variants of interest and concern. Frank Konings, Mark D. Perkins, Jens H. Kuhn, Mark J. Pallen, Erik J. Alm, Brett N. Archer, Amal Barakat, Trevor Bedford, Jinal N. Bhiman, Leon Caly, Lisa L. Carter, Anne Cullinane, Tulio de Oliveira, Julian Druce, Ihab El Masry, Roger Evans, George F. Gao, Alexander E. Gorbalenya, Esther Hamblion, Belinda L. Herring, Emma Hodcroft, Edward C. Holmes, Manish Kakkar, Shagun Khare, Marion P. G. Koopmans, Bette Korber, Juliana Leite, Duncan MacCannell, Marco Marklewitz, Sebastian Maurer-Stroh, Jairo Andres Mendez Rico, Vincent J. Munster, Richard Neher, Bas Oude Munnink, Boris I. Pavlin, Malik Peiris, Leo Poon, Oliver Pybus, Andrew Rambaut, Paola Resende, Lorenzo Subissi, Volker Thiel, Suxiang Tong, Sylvie van der Werf, Anne von Gottberg, John Ziebuhr and Maria D. Van Kerkhove evere acute respiratory syndrome The WHO has provided working the same time and without regard for coronavirus 2 (SARS-CoV-2), the definitions of variants of interest (VOIs) the properties of VOIs and VOCs. causative agent of coronavirus and variants of concern (VOCs) that will The lack of clarity on naming VOCs S 2 disease 2019 (COVID-19), has a linear, be updated when required . These working makes it difficult for those who are not unsegmented, positive-sense RNA definitions also consider changes that lead experts in the field to link such variants genome. -

The PHA4GE SARS-Cov-2 Contextual Data Specification for Open Genomic Epidemiology
Preprints (www.preprints.org) | NOT PEER-REVIEWED | Posted: 9 August 2020 doi:10.20944/preprints202008.0220.v1 The PHA4GE SARS-CoV-2 contextual data specification for open genomic epidemiology Authors 1* 2 3 3 Emma J Griffiths , Ruth E Timme , Andrew J Page , Nabil-Fareed Alikhan , Dan 4 5 6 7 8 Fornika , Finlay Maguire , Catarina Inês Mendes , Simon H Tausch , Allison Black , 9,10 11 12, 13 14 Thomas R Connor , Gregory H Tyson , David M Aanensen , Brian Alcock , 15 16 17 Josefina Campos , Alan Christoffels , Anders Gonçalves da Silva , Emma Hodcroft 18 1, 19, 20 21, 22 23, 24 , William WL Hsiao , Lee S Katz , Samuel M Nicholls , Paul E Oluniyi , 23, 24 14 25 Idowu B Olawoye , Amogelang R Raphenya , Ana Tereza R Vasconcelos , Adam 26 27 A Witney , and Duncan R MacCannell , on behalf of the Public Health Alliance for Genomic Epidemiology (PHA4GE) consortium. Affiliations 1 Department of Pathology and Laboratory Medicine, University of British Columbia, Vancouver, British Columbia, Canada 2 Center for Food Safety and Applied Nutrition, U.S. Food and Drug Administration, College Park, MD, USA. 3 Quadram Institute Bioscience, Norwich, Norfolk, UK 4 BC Centre for Disease Control Public Health Laboratory, Vancouver, Canada 5 Faculty of Computer Science, Dalhousie University, Halifax, Canada 6 Instituto de Microbiologia, Instituto de Medicina Molecular, Faculdade de Medicina, Universidade de Lisboa, Lisboa, Portugal 7 Department of Biological Safety, German Federal Institute for Risk Assessment, Berlin, Germany 8 Department of Epidemiology, University of Washington, Washington, USA. 9 Organisms and Environment Division, School of Biosciences, Cardiff University, Cardiff, Wales, UK. -

World Service Listings for 31 October – 6
World Service Listings for 31 October – 6 November 2020 Page 1 of 15 SATURDAY 31 OCTOBER 2020 A BBC Arabic investigation has uncovered systemic child abuse embracing the iconic film character, Borat! inside Islamic schools in Sudan, known as khalwas. It found that SAT 00:00 BBC News (w172x5p439m5t3l) boys are routinely chained, shackled and beaten; in some Joining Julian Worricker to discuss these and other issues: The latest five minute news bulletin from BBC World Service. khalwas there was evidence of sexual abuse. The BBC’s Azadeh Moaveni, Iranian author and Crisis Group's Gender Mamdouh Akbiek worked with a local investigative journalist Project Director, and James Rodgers, writer and lecturer in on the story. international journalism. SAT 00:06 The Real Story (w3cszcns) US v China: A new Cold War? Boston's Brazilian party-goers (Photo:Rescuers and local residents are frantically searching for Boston’s large Brazilian community has stirred up controversy survivors in Izmir. Credit: Reuters) The central committee of China’s ruling Communist Party has by holding huge outdoor parties despite Covid-19 restrictions, been meeting this week in Beijing to map out its priorities for as BBC Brasil’s Ricardo Senra explains. the next five years. While Americans decide whether Donald SAT 07:00 BBC News (w172x5p439m6nbh) Trump or Joe Biden will set the direction of US foreign policy Somali storm in a teacup News bulletin going forward, there is little doubt that Chinese President Xi A social media star was deported from the self-declared Jinping will remain in his post for the foreseeable future - party republic of Somaliland because of a cup of tea. -

NGS-Final-Program-2020.Pdf
ASM Conference on Rapid Applied Microbial Next- Generation Sequencing and Bioinformatic Pipelines December 7-11, 2020 • Online Final Program See sequencing in a new light Explore the NextSeq™ 2000 www.illumina.com/NextSeq2000 For Research Use Only. Not for use in diagnostic procedures. © 2020 Illumina, Inc. All rights reserved. TABLE OF CONTENTS Program Committee .........................................................................2 Acknowledgments .............................................................................2 General Information .........................................................................3 Program ..............................................................................................5 Facebook: facebook.com/asmfan Follow ASM Twitter: twitter.com/asmicrobiology LinkedIn: linkedin.com/company/american- society-for-microbiology Instagram: instagram.com/asmicrobiology YouTube: http://bit.ly/ASMyoutubesub Join the conversation on social media and use hashtag #ASMNGS! 1 PROGRAM COMMITTEE Duncan MacCannell - Jennifer Gardy Paul Keim Chair Bill & Melinda Gates Northern Arizona Centers for Disease Foundation University and TGen Control and Prevention, Seattle, Wash. North, Flagstaff, Ariz. Atlanta, Ga. Elodie Ghedin Adam Phillippy Ruth Timme - Co-Chair New York University National Institutes of U.S. Food and Drug New York, N.Y. Health, Bethesda, Md. Administration College Park, Md. George Gthinji Torsten Seemann KEMRI/Wellcome Trust University of Melbourne Marc Allard Nairobi, Kenya Melbourne, Australia -

Impact Assessment Feedback (2 of 3)
Deliverable D9.5b Impact Assessment Feedback (2 of 3) This deliverable is part of a project that has received funding from the European Union’s Horizon 2020 research and innovation programme Grant Agreement No: 770523 Project dates: 1 December 2017 - 30 November 2021 www.InsSciDE.eu InsSciDE has received funding under the European Union's Horizon 2020 research and innovation program (Grant Agreement No 770523, 2018-2021) To be cited as: Palmberg, D., Mays, C., Bürgi Dellsperger, C. “Beyond Quantitative: InsSciDE Project Impact Assessment Feedback”. Deliverable n° 9.5b H2020 InsSciDE project, submitted by UNESCO, November, 2020. This report was prepared under Grant Agreement n° 770523 from the European Commission. The content of this document does not necessarily reflect the official opinions of the European Commission or other institution of the European Union. Deliverable title: Impact Assessment Feedback Deliverable n°: 9.5b Nature of the deliverable: Report Dissemination level: Confidential (Co) Lead beneficiary: UNESCO – partner n°4 Lead author: Daniella Palmberg, UNESCO, [email protected] Contributing authors: Claire Mays, Christina Bürgi Dellsperger, Rasmus Bertelsen, David Burigana, Miyase Christensen, Maria Paula Diogo, Katharina Paul, Alexander Pruss, Simone Turchetti, Nina Wormbs Reviewed by: Claire Mays, SYMLOG; Leonard Laborie, CNRS Approved by: Christina Bürgi Dellsperger, UNESCO Due date of deliverable: Month n° 35 Submitted to coordinator: Month n° 38 ABSTRACT InsSciDE (Inventing a shared Science Diplomacy for Europe) seeks to engage historians of science and technology, networks of diplomats and scientists, and experts of strategy and policy makers, to increase understanding of science diplomacy and offer frameworks and guidelines for its use. -

The John Snow Memorandum (PDF)
MORE THAN 2,000 scientists, researchers & healthcare professionals have now signed the John Snow Memorandum. THE JOHN SNOW MEMORANDUM Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has infected more than 35 million people globally, with more than 1 million deaths recorded by the World Health Organization as of Oct 12, 2020. As a second wave of COVID-19 affects Europe, and with winter approaching, we need clear communication about the risks posed by COVID-19 and effective strategies to combat them. Here, we share our view of the current evidence-based consensus on COVID-19. The John Snow Memorandum October 14, 2020 SARS-CoV-2 spreads through contact (via larger droplets and aerosols), and longer- range transmission via aerosols, especially in conditions where ventilation is poor. Its high infectivity(1) combined with the susceptibility of unexposed populations to a new virus, creates conditions for rapid community spread. The infection fatality rate of COVID-19 is several-fold higher than that of seasonal influenza(2) and infection can lead to persisting illness, including in young, previously healthy people (ie, long COVID(3)). It is unclear how long protective immunity lasts(4) and, like other seasonal coronaviruses, SARS-CoV-2 is capable of re-infecting people who have already had the disease, but the frequency of re-infection is unknown(5). Transmission of the virus can be mitigated through physical distancing, use of face coverings, hand and respiratory hygiene, and by avoiding crowds and poorly ventilated spaces. Rapid testing, contact tracing, and isolation are also critical to controlling transmission. The World Health Organization has been advocating for these measures since early in the pandemic. -

Building a New Biology
BUILDING A NEW BIOLOGY School of Biological Sciences 2025 Vision BUILDING A Contents NEW BIOLOGY School of Biological Sciences 2025 Vision Foreword 04 Executive summary 05 CHAPTER 1: Vision for the future 06 1:1 Our School 08 1:2 World-class research 10 1:3 Inspirational teaching 14 1:4 Creative communication 15 1:5 Pioneering innovation 16 1:6 Economic benefits 18 1:7 Global impact 19 CHAPTER 2: Building a new biology 22 2:1 Epigenetics 24 2:2 Infection and global health 25 2:3 Synthetic biology 26 2:4 At the interface 27 CHAPTER 3: A blueprint for growth 28 3:1 Our people 30 3:2 Our culture 33 3:3 Our spaces 34 3:4 Our future impact 36 Conclusion 37 www.ed.ac.uk/schools-departments/biology Foreword Executive summary 1.1 We are poised to deliver We have embarked on an estate re-development project that will The School of Biological Sciences at the University of Edinburgh is Our goals the next revolution in see enhanced integration of our research activities, nurturing the a leading centre for research and teaching with an enviable global biology. Today, biology cross disciplinary collaborations. Some of the exceptional research reach and reputation. For centuries we have generated insights and We have an ambitious long-term vision for our School and by is no longer simply that will arise from novel co-locations in our new buildings can innovations that have benefited the health and wealth of our world. 2025 we aim to: about interrogation and be foreseen, for instance just by browsing the pages here.