Distribution, Interaction and Functional Profiles of Epiphytic Bacterial
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Influence of Probiotics on the Firmicutes/Bacteroidetes Ratio In
microorganisms Review The Influence of Probiotics on the Firmicutes/Bacteroidetes Ratio in the Treatment of Obesity and Inflammatory Bowel disease Spase Stojanov 1,2, Aleš Berlec 1,2 and Borut Štrukelj 1,2,* 1 Faculty of Pharmacy, University of Ljubljana, SI-1000 Ljubljana, Slovenia; [email protected] (S.S.); [email protected] (A.B.) 2 Department of Biotechnology, Jožef Stefan Institute, SI-1000 Ljubljana, Slovenia * Correspondence: borut.strukelj@ffa.uni-lj.si Received: 16 September 2020; Accepted: 31 October 2020; Published: 1 November 2020 Abstract: The two most important bacterial phyla in the gastrointestinal tract, Firmicutes and Bacteroidetes, have gained much attention in recent years. The Firmicutes/Bacteroidetes (F/B) ratio is widely accepted to have an important influence in maintaining normal intestinal homeostasis. Increased or decreased F/B ratio is regarded as dysbiosis, whereby the former is usually observed with obesity, and the latter with inflammatory bowel disease (IBD). Probiotics as live microorganisms can confer health benefits to the host when administered in adequate amounts. There is considerable evidence of their nutritional and immunosuppressive properties including reports that elucidate the association of probiotics with the F/B ratio, obesity, and IBD. Orally administered probiotics can contribute to the restoration of dysbiotic microbiota and to the prevention of obesity or IBD. However, as the effects of different probiotics on the F/B ratio differ, selecting the appropriate species or mixture is crucial. The most commonly tested probiotics for modifying the F/B ratio and treating obesity and IBD are from the genus Lactobacillus. In this paper, we review the effects of probiotics on the F/B ratio that lead to weight loss or immunosuppression. -

Early Photosynthetic Eukaryotes Inhabited Low-Salinity Habitats
Early photosynthetic eukaryotes inhabited PNAS PLUS low-salinity habitats Patricia Sánchez-Baracaldoa,1, John A. Ravenb,c, Davide Pisanid,e, and Andrew H. Knollf aSchool of Geographical Sciences, University of Bristol, Bristol BS8 1SS, United Kingdom; bDivision of Plant Science, University of Dundee at the James Hutton Institute, Dundee DD2 5DA, United Kingdom; cPlant Functional Biology and Climate Change Cluster, University of Technology Sydney, Ultimo, NSW 2007, Australia; dSchool of Biological Sciences, University of Bristol, Bristol BS8 1TH, United Kingdom; eSchool of Earth Sciences, University of Bristol, Bristol BS8 1TH, United Kingdom; and fDepartment of Organismic and Evolutionary Biology, Harvard University, Cambridge, MA 02138 Edited by Peter R. Crane, Oak Spring Garden Foundation, Upperville, Virginia, and approved July 7, 2017 (received for review December 7, 2016) The early evolutionary history of the chloroplast lineage remains estimates for the origin of plastids ranging over 800 My (7). At the an open question. It is widely accepted that the endosymbiosis that same time, the ecological setting in which this endosymbiotic event established the chloroplast lineage in eukaryotes can be traced occurred has not been fully explored (8), partly because of phy- back to a single event, in which a cyanobacterium was incorpo- logenetic uncertainties and preservational biases of the fossil re- rated into a protistan host. It is still unclear, however, which cord. Phylogenomics and trait evolution analysis have pointed to a Cyanobacteria are most closely related to the chloroplast, when the freshwater origin for Cyanobacteria (9–11), providing an approach plastid lineage first evolved, and in what habitats this endosym- to address the early diversification of terrestrial biota for which the biotic event occurred. -
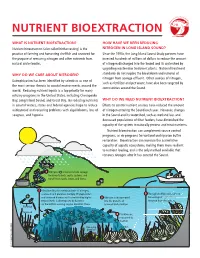
Nutrient Bioextraction L
soun nd d s sla tu i d g y n o NUTRIENT BIOEXTRACTION l WHAT IS NUTRIENT BIOEXTRACTION? HOW HAVE WE BEEN REDUCING Nutrient bioextraction (also called bioharvesting) is the NITROGEN IN LONG ISLAND SOUND? practice of farming and harvesting shellfish and seaweed for Since the 1990s, the Long Island Sound Study partners have the purpose of removing nitrogen and other nutrients from invested hundreds of millions of dollars to reduce the amount natural water bodies. of nitrogen discharged into the Sound and its watershed by upgrading wastewater treatment plants. National treatment WHY DO WE CARE ABOUT NITROGEN? standards do not require the breakdown and removal of nitrogen from sewage effluent. Other sources of nitrogen, Eutrophication has been identified by scientists as one of such as fertilizer and pet waste, have also been targeted by the most serious threats to coastal environments around the communities around the Sound. world. Reducing nutrient inputs is a top priority for many estuary programs in the United States, including Chesapeake Bay, Long Island Sound, and Great Bay. By reducing nutrients WHY DO WE NEED NUTRIENT BIOEXTRACTION? in coastal waters, states and federal agencies hope to reduce Efforts to control nutrient sources have reduced the amount widespread and recurring problems with algal blooms, loss of of nitrogen entering the Sound each year. However, changes seagrass, and hypoxia. in the Sound and its watershed, such as wetland loss and decreased populations of filter feeders, have diminished the capacity of the system to naturally process and treat nutrients. Nutrient bioextraction can complement source control programs, as do programs for wetland and riparian buffer restoration. -

High-Quality Draft Genome Sequences of Five Anaerobic Oral Bacteria and Description of Peptoanaerobacter Stomatis Gen
Sizova et al. Standards in Genomic Sciences (2015) 10:37 DOI 10.1186/s40793-015-0027-8 EXTENDED GENOME REPORT Open Access High-quality draft genome sequences of five anaerobic oral bacteria and description of Peptoanaerobacter stomatis gen. nov., sp. nov., a new member of the family Peptostreptococcaceae Maria V. Sizova1, Amanda Chilaka1, Ashlee M. Earl2, Sebastian N. Doerfert1, Paul A. Muller1, Manolito Torralba3, Jamison M. McCorrison3, A. Scott Durkin3, Karen E. Nelson3 and Slava S. Epstein1* Abstract Here we report a summary classification and the features of five anaerobic oral bacteria from the family Peptostreptococcaceae. Bacterial strains were isolated from human subgingival plaque. Strains ACC19a, CM2, CM5, and OBRC8 represent the first known cultivable members of “yet uncultured” human oral taxon 081; strain AS15 belongs to “cultivable” human oral taxon 377. Based on 16S rRNA gene sequence comparisons, strains ACC19a, CM2, CM5, and OBRC8 are distantly related to Eubacterium yurii subs. yurii and Filifactor alocis, with 93.2 – 94.4 % and 85.5 % of sequence identity, respectively. The genomes of strains ACC19a, CM2, CM5, OBRC8 and AS15 are 2,541,543; 2,312,592; 2,594,242; 2,553,276; and 2,654,638 bp long. The genomes are comprised of 2277, 1973, 2325, 2277, and 2308 protein-coding genes and 54, 57, 54, 36, and 28 RNA genes, respectively. Based on the distinct characteristics presented here, we suggest that strains ACC19a, CM2, CM5, and OBRC8 represent a novel genus and species within the family Peptostreptococcaceae, for which we propose the name Peptoanaerobacter stomatis gen. nov., sp. nov. The type strain is strain ACC19aT (=HM-483T; =DSM 28705T;=ATCC BAA-2665T). -

Genomics 98 (2011) 370–375
Genomics 98 (2011) 370–375 Contents lists available at ScienceDirect Genomics journal homepage: www.elsevier.com/locate/ygeno Whole-genome comparison clarifies close phylogenetic relationships between the phyla Dictyoglomi and Thermotogae Hiromi Nishida a,⁎, Teruhiko Beppu b, Kenji Ueda b a Agricultural Bioinformatics Research Unit, Graduate School of Agricultural and Life Sciences, University of Tokyo, 1-1-1 Yayoi, Bunkyo-ku, Tokyo 113-8657, Japan b Life Science Research Center, College of Bioresource Sciences, Nihon University, Fujisawa, Japan article info abstract Article history: The anaerobic thermophilic bacterial genus Dictyoglomus is characterized by the ability to produce useful Received 2 June 2011 enzymes such as amylase, mannanase, and xylanase. Despite the significance, the phylogenetic position of Accepted 1 August 2011 Dictyoglomus has not yet been clarified, since it exhibits ambiguous phylogenetic positions in a single gene Available online 7 August 2011 sequence comparison-based analysis. The number of substitutions at the diverging point of Dictyoglomus is insufficient to show the relationships in a single gene comparison-based analysis. Hence, we studied its Keywords: evolutionary trait based on whole-genome comparison. Both gene content and orthologous protein sequence Whole-genome comparison Dictyoglomus comparisons indicated that Dictyoglomus is most closely related to the phylum Thermotogae and it forms a Bacterial systematics monophyletic group with Coprothermobacter proteolyticus (a constituent of the phylum Firmicutes) and Coprothermobacter proteolyticus Thermotogae. Our findings indicate that C. proteolyticus does not belong to the phylum Firmicutes and that the Thermotogae phylum Dictyoglomi is not closely related to either the phylum Firmicutes or Synergistetes but to the phylum Thermotogae. © 2011 Elsevier Inc. -

Detection of a Bacterial Group Within the Phylum Chloroflexi And
Microbes Environ. Vol. 21, No. 3, 154–162, 2006 http://wwwsoc.nii.ac.jp/jsme2/ Detection of a Bacterial Group within the Phylum Chloroflexi and Reductive-Dehalogenase-Homologous Genes in Pentachlorobenzene- Dechlorinating Estuarine Sediment from the Arakawa River, Japan KYOSUKE SANTOH1, ATSUSHI KOUZUMA1, RYOKO ISHIZEKI2, KENICHI IWATA1, MINORU SHIMURA3, TOSHIO HAYAKAWA3, TOSHIHIRO HOAKI4, HIDEAKI NOJIRI1, TOSHIO OMORI2, HISAKAZU YAMANE1 and HIROSHI HABE1*† 1 Biotechnology Research Center, The University of Tokyo, 1–1–1 Yayoi, Bunkyo-ku, Tokyo 113–8657, Japan 2 Department of Industrial Chemistry, Faculty of Engineering, Shibaura Institute of Technology, Minato-ku, Tokyo 108–8548, Japan 3 Environmental Biotechnology Laboratory, Railway Technical Research Institute, 2–8–38 Hikari-cho, Kokubunji-shi, Tokyo 185–8540, Japan 4 Technology Research Center, Taisei Corporation, 344–1 Nase, Totsuka-ku, Yokohama 245–0051, Japan (Received April 21, 2006—Accepted June 12, 2006) We enriched a pentachlorobenzene (pentaCB)-dechlorinating microbial consortium from an estuarine-sedi- ment sample obtained from the mouth of the Arakawa River. The sediment was incubated together with a mix- ture of four electron donors and pentaCB, and after five months of incubation, the microbial community structure was analyzed. Both DGGE and clone library analyses showed that the most expansive phylogenetic group within the consortium was affiliated with the phylum Chloroflexi, which includes Dehalococcoides-like bacteria. PCR using a degenerate primer set targeting conserved regions in reductive-dehalogenase-homologous (rdh) genes from Dehalococcoides species revealed that DNA fragments (approximately 1.5–1.7 kb) of rdh genes were am- plified from genomic DNA of the consortium. The deduced amino acid sequences of the rdh genes shared sever- al characteristics of reductive dehalogenases. -

Algae & Marine Plants of Point Reyes
Algae & Marine Plants of Point Reyes Green Algae or Chlorophyta Genus/Species Common Name Acrosiphonia coalita Green rope, Tangled weed Blidingia minima Blidingia minima var. vexata Dwarf sea hair Bryopsis corticulans Cladophora columbiana Green tuft alga Codium fragile subsp. californicum Sea staghorn Codium setchellii Smooth spongy cushion, Green spongy cushion Trentepohlia aurea Ulva californica Ulva fenestrata Sea lettuce Ulva intestinalis Sea hair, Sea lettuce, Gutweed, Grass kelp Ulva linza Ulva taeniata Urospora sp. Brown Algae or Ochrophyta Genus/Species Common Name Alaria marginata Ribbon kelp, Winged kelp Analipus japonicus Fir branch seaweed, Sea fir Coilodesme californica Dactylosiphon bullosus Desmarestia herbacea Desmarestia latifrons Egregia menziesii Feather boa Fucus distichus Bladderwrack, Rockweed Haplogloia andersonii Anderson's gooey brown Laminaria setchellii Southern stiff-stiped kelp Laminaria sinclairii Leathesia marina Sea cauliflower Melanosiphon intestinalis Twisted sea tubes Nereocystis luetkeana Bull kelp, Bullwhip kelp, Bladder wrack, Edible kelp, Ribbon kelp Pelvetiopsis limitata Petalonia fascia False kelp Petrospongium rugosum Phaeostrophion irregulare Sand-scoured false kelp Pterygophora californica Woody-stemmed kelp, Stalked kelp, Walking kelp Ralfsia sp. Silvetia compressa Rockweed Stephanocystis osmundacea Page 1 of 4 Red Algae or Rhodophyta Genus/Species Common Name Ahnfeltia fastigiata Bushy Ahnfelt's seaweed Ahnfeltiopsis linearis Anisocladella pacifica Bangia sp. Bossiella dichotoma Bossiella -

Fatty Acid Diets: Regulation of Gut Microbiota Composition and Obesity and Its Related Metabolic Dysbiosis
International Journal of Molecular Sciences Review Fatty Acid Diets: Regulation of Gut Microbiota Composition and Obesity and Its Related Metabolic Dysbiosis David Johane Machate 1, Priscila Silva Figueiredo 2 , Gabriela Marcelino 2 , Rita de Cássia Avellaneda Guimarães 2,*, Priscila Aiko Hiane 2 , Danielle Bogo 2, Verônica Assalin Zorgetto Pinheiro 2, Lincoln Carlos Silva de Oliveira 3 and Arnildo Pott 1 1 Graduate Program in Biotechnology and Biodiversity in the Central-West Region of Brazil, Federal University of Mato Grosso do Sul, Campo Grande 79079-900, Brazil; [email protected] (D.J.M.); [email protected] (A.P.) 2 Graduate Program in Health and Development in the Central-West Region of Brazil, Federal University of Mato Grosso do Sul, Campo Grande 79079-900, Brazil; pri.fi[email protected] (P.S.F.); [email protected] (G.M.); [email protected] (P.A.H.); [email protected] (D.B.); [email protected] (V.A.Z.P.) 3 Chemistry Institute, Federal University of Mato Grosso do Sul, Campo Grande 79079-900, Brazil; [email protected] * Correspondence: [email protected]; Tel.: +55-67-3345-7416 Received: 9 March 2020; Accepted: 27 March 2020; Published: 8 June 2020 Abstract: Long-term high-fat dietary intake plays a crucial role in the composition of gut microbiota in animal models and human subjects, which affect directly short-chain fatty acid (SCFA) production and host health. This review aims to highlight the interplay of fatty acid (FA) intake and gut microbiota composition and its interaction with hosts in health promotion and obesity prevention and its related metabolic dysbiosis. -

Microbial Life Under Ice: Metagenome Diversity and in Situ Activity Of
bioRxiv preprint doi: https://doi.org/10.1101/324970; this version posted May 17, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Microbial life under ice: metagenome diversity and in situ activity of Verrucomicrobia in seasonally ice-covered lakes Patricia Tran1,2, Arthi Ramachandran1, Ola Khawasik1, Beatrix E. Beisner2,3, Milla Rautio2,4, Yannick Huot,2,5, David A. Walsh1,2 1 Department of Biology, Concordia University, 7141 Sherbrooke St. West, Montreal, Quebec, H4B 1R6, Canada 2 Groupe de recherche interuniversitaire en limnologie et environnement aquatique (GRIL), Montréal, Québec, Canada 3 Département des sciences biologiques, Université du Québec à Montréal, Québec, Canada. 4 Département des sciences fondamentales, Université du Québec à Chicoutimi, Chicoutimi, Québec, Canada 5 Département de géomatique appliquée, Université de Sherbrooke, Sherbrooke, Québec, Canada Corresponding author: David Walsh, Department of Biology, Concordia University, 7141 Sherbrooke St. West Montreal, QC H4B 1R6 Canada 514-848-2424 ext 3477 [email protected] Running title: Sub-ice Verrucomicrobia genomes in Quebec lakes bioRxiv preprint doi: https://doi.org/10.1101/324970; this version posted May 17, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 Summary 2 Northern lakes are ice-covered for a large part of the year, yet our understanding 3 of microbial diversity and activity during winter lags behind that of the ice-free period. In 4 this study, we investigated under-ice diversity and metabolism of Verrucomicrobia in 5 seasonally ice-covered lakes in temperate and boreal regions of Quebec, Canada using 6 16S rRNA sequencing, metagenomics and metatranscriptomics. -

Effect of Ph and Temperature on Microbial Community Structure And
Calicioglu et al. Biotechnol Biofuels (2018) 11:275 https://doi.org/10.1186/s13068-018-1278-6 Biotechnology for Biofuels RESEARCH Open Access Efect of pH and temperature on microbial community structure and carboxylic acid yield during the acidogenic digestion of duckweed Ozgul Calicioglu1, Michael J. Shreve1, Tom L. Richard2 and Rachel A. Brennan1* Abstract Background: Duckweeds (Lemnaceae) are efcient aquatic plants for wastewater treatment due to their high nutri- ent-uptake capabilities and resilience to severe environmental conditions. Combined with their rapid growth rates, high starch, and low lignin contents, duckweeds have also gained popularity as a biofuel feedstock for thermochemi- cal conversion and alcohol fermentation. However, studies on the acidogenic anaerobic digestion of duckweed into carboxylic acids, another group of chemicals which are precursors of higher-value chemicals and biofuels, are lacking. In this study, a series of laboratory batch experiments were performed to determine the favorable operating condi- tions (i.e., temperature and pH) to maximize carboxylic acid production from wastewater-derived duckweed during acidogenic digestion. Batch reactors with 25 g/l solid loading were operated anaerobically for 21 days under meso- philic (35 °C) or thermophilic (55 °C) conditions at an acidic (5.3) or basic (9.2) pH. At the conclusion of the experiment, the dominant microbial communities under various operating conditions were assessed using high-throughput sequencing. Results: The highest duckweed–carboxylic acid conversion of 388 28 mg acetic acid equivalent per gram volatile solids was observed under mesophilic and basic conditions, with an± average production rate of 0.59 g/l/day. This result is comparable to those reported for acidogenic digestion of other organics such as food waste. -

Kelp Cultivation Effectively Improves Water Quality and Regulates Phytoplankton Community in a Turbid, Highly Eutrophic Bay
Science of the Total Environment 707 (2020) 135561 Contents lists available at ScienceDirect Science of the Total Environment journal homepage: www.elsevier.com/locate/scitotenv Kelp cultivation effectively improves water quality and regulates phytoplankton community in a turbid, highly eutrophic bay Zhibing Jiang a,b,d, Jingjing Liu a,ShangluLic,YueChena,PingDua,b,YuanliZhua,YiboLiaoa,d, Quanzhen Chen a, Lu Shou a, Xiaojun Yan e, Jiangning Zeng a,⁎, Jianfang Chen a,d a Key Laboratory of Marine Ecosystem and Biogeochemistry, State Oceanic Administration & Second Institute of Oceanography, Ministry of Natural Resources, Hangzhou, China b Function Laboratory for Marine Fisheries Science and Food Production Processes, Pilot National Laboratory for Marine Science and Technology (Qingdao), Qingdao, China c Marine Monitoring and Forecasting Center of Zhejiang Province, Hangzhou, China d State Key Laboratory of Satellite Ocean Environment Dynamics, Second Institute of Oceanography, Ministry of Natural Resources, Hangzhou, China e Key Laboratory of Applied Marine Biotechnology, Ministry of Education, Marine College of Ningbo University, Ningbo, China HIGHLIGHTS GRAPHICAL ABSTRACT • Kelp farming alleviated eutrophication and acidification. • Kelp farming greatly relieved light limi- tation and increased phytoplankton bio- mass. • Kelp farming appreciably enhanced phytoplankton diversity. • Kelp farming reduced the dominance of dinoflagellate Prorocentrum minimum. • Phytoplankton community differed sig- nificantly between the kelp farm and control area. article info abstract Article history: Coastal eutrophication and its associated harmful algal blooms have emerged as one of the most severe environ- Received 19 September 2019 mental problems worldwide. Seaweed cultivation has been widely encouraged to control eutrophication and Received in revised form 11 November 2019 algal blooms. Among them, cultivated kelp (Saccharina japonica) dominates primarily by production and area. -

Cone-Forming Chloroflexi Mats As Analogs of Conical
268 Appendix 2 CONE-FORMING CHLOROFLEXI MATS AS ANALOGS OF CONICAL STROMATOLITE FORMATION WITHOUT CYANOBACTERIA Lewis M. Ward, Woodward W. Fischer, Katsumi Matsuura, and Shawn E. McGlynn. In preparation. Abstract Modern microbial mats provide useful process analogs for understanding the mechanics behind the production of ancient stromatolites. However, studies to date have focused on mats composed predominantly of oxygenic Cyanobacteria (Oxyphotobacteria) and algae, which makes it difficult to assess a unique role of oxygenic photosynthesis in stromatolite morphogenesis, versus different mechanics such as phototaxis and filamentous growth. Here, we characterize Chloroflexi-rich hot spring microbial mats from Nakabusa Onsen, Nagano Prefecture, Japan. This spring supports cone-forming microbial mats in both upstream high-temperature, sulfidic regions dominated by filamentous anoxygenic phototrophic Chloroflexi, as well as downstream Cyanobacteria-dominated mats. These mats produce similar morphologies analogous to conical stromatolites despite metabolically and taxonomically divergent microbial communities as revealed by 16S and shotgun metagenomic sequencing and microscopy. These data illustrate that anoxygenic filamentous microorganisms appear to be capable of producing similar mat morphologies as those seen in Oxyphotobacteria-dominated systems and commonly associated with 269 conical Precambrian stromatolites, and that the processes leading to the development of these features is more closely related with characteristics such as hydrology and cell morphology and motility. Introduction Stromatolites are “attached, lithified sedimentary growth structures, accretionary away from a point or limited surface of initiation” (Grotzinger and Knoll 1999). Behind this description lies a wealth of sedimentary structures with a record dating back over 3.7 billion years that may be one of the earliest indicators of life on Earth (Awramik 1992, Nutman et al.