Phylogenetic Diversity of Sediment Bacteria from the Deep Northeastern Pacific Ocean: a Comparison with the Deep Eastern Mediterranean Sea
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Legionella Shows a Diverse Secondary Metabolism Dependent on a Broad Spectrum Sfp-Type Phosphopantetheinyl Transferase
Legionella shows a diverse secondary metabolism dependent on a broad spectrum Sfp-type phosphopantetheinyl transferase Nicholas J. Tobias1, Tilman Ahrendt1, Ursula Schell2, Melissa Miltenberger1, Hubert Hilbi2,3 and Helge B. Bode1,4 1 Fachbereich Biowissenschaften, Merck Stiftungsprofessur fu¨r Molekulare Biotechnologie, Goethe Universita¨t, Frankfurt am Main, Germany 2 Max von Pettenkofer Institute, Ludwig-Maximilians-Universita¨tMu¨nchen, Munich, Germany 3 Institute of Medical Microbiology, University of Zu¨rich, Zu¨rich, Switzerland 4 Buchmann Institute for Molecular Life Sciences, Goethe Universita¨t, Frankfurt am Main, Germany ABSTRACT Several members of the genus Legionella cause Legionnaires’ disease, a potentially debilitating form of pneumonia. Studies frequently focus on the abundant number of virulence factors present in this genus. However, what is often overlooked is the role of secondary metabolites from Legionella. Following whole genome sequencing, we assembled and annotated the Legionella parisiensis DSM 19216 genome. Together with 14 other members of the Legionella, we performed comparative genomics and analysed the secondary metabolite potential of each strain. We found that Legionella contains a huge variety of biosynthetic gene clusters (BGCs) that are potentially making a significant number of novel natural products with undefined function. Surprisingly, only a single Sfp-like phosphopantetheinyl transferase is found in all Legionella strains analyzed that might be responsible for the activation of all carrier proteins in primary (fatty acid biosynthesis) and secondary metabolism (polyketide and non-ribosomal peptide synthesis). Using conserved active site motifs, we predict Submitted 29 June 2016 some novel compounds that are probably involved in cell-cell communication, Accepted 25 October 2016 Published 24 November 2016 differing to known communication systems. -

Characterization of Arsenite-Oxidizing Bacteria Isolated from Arsenic-Rich Sediments, Atacama Desert, Chile
microorganisms Article Characterization of Arsenite-Oxidizing Bacteria Isolated from Arsenic-Rich Sediments, Atacama Desert, Chile Constanza Herrera 1, Ruben Moraga 2,*, Brian Bustamante 1, Claudia Vilo 1, Paulina Aguayo 1,3,4, Cristian Valenzuela 1, Carlos T. Smith 1 , Jorge Yáñez 5, Victor Guzmán-Fierro 6, Marlene Roeckel 6 and Víctor L. Campos 1,* 1 Laboratory of Environmental Microbiology, Department of Microbiology, Faculty of Biological Sciences, Universidad de Concepcion, Concepcion 4070386, Chile; [email protected] (C.H.); [email protected] (B.B.); [email protected] (C.V.); [email protected] (P.A.); [email protected] (C.V.); [email protected] (C.T.S.) 2 Microbiology Laboratory, Faculty of Renewable Natural Resources, Arturo Prat University, Iquique 1100000, Chile 3 Faculty of Environmental Sciences, EULA-Chile, Universidad de Concepcion, Concepcion 4070386, Chile 4 Institute of Natural Resources, Faculty of Veterinary Medicine and Agronomy, Universidad de Las Américas, Sede Concepcion, Campus El Boldal, Av. Alessandri N◦1160, Concepcion 4090940, Chile 5 Faculty of Chemical Sciences, Department of Analytical and Inorganic Chemistry, University of Concepción, Concepción 4070386, Chile; [email protected] 6 Department of Chemical Engineering, Faculty of Engineering, University of Concepción, Concepcion 4070386, Chile; victorguzmanfi[email protected] (V.G.-F.); [email protected] (M.R.) * Correspondence: [email protected] (R.M.); [email protected] (V.L.C.) Abstract: Arsenic (As), a semimetal toxic for humans, is commonly associated -

The Risk to Human Health from Free-Living Amoebae Interaction with Legionella in Drinking and Recycled Water Systems
THE RISK TO HUMAN HEALTH FROM FREE-LIVING AMOEBAE INTERACTION WITH LEGIONELLA IN DRINKING AND RECYCLED WATER SYSTEMS Dissertation submitted by JACQUELINE MARIE THOMAS BACHELOR OF SCIENCE (HONOURS) AND BACHELOR OF ARTS, UNSW In partial fulfillment of the requirements for the award of DOCTOR OF PHILOSOPHY in ENVIRONMENTAL ENGINEERING SCHOOL OF CIVIL AND ENVIRONMENTAL ENGINEERING FACULTY OF ENGINEERING MAY 2012 SUPERVISORS Professor Nicholas Ashbolt Office of Research and Development United States Environmental Protection Agency Cincinnati, Ohio USA and School of Civil and Environmental Engineering Faculty of Engineering The University of New South Wales Sydney, Australia Professor Richard Stuetz School of Civil and Environmental Engineering Faculty of Engineering The University of New South Wales Sydney, Australia Doctor Torsten Thomas School of Biotechnology and Biomolecular Sciences Faculty of Science The University of New South Wales Sydney, Australia ORIGINALITY STATEMENT '1 hereby declare that this submission is my own work and to the best of my knowledge it contains no materials previously published or written by another person, or substantial proportions of material which have been accepted for the award of any other degree or diploma at UNSW or any other educational institution, except where due acknowledgement is made in the thesis. Any contribution made to the research by others, with whom 1 have worked at UNSW or elsewhere, is explicitly acknowledged in the thesis. I also declare that the intellectual content of this thesis is the product of my own work, except to the extent that assistance from others in the project's design and conception or in style, presentation and linguistic expression is acknowledged.' Signed ~ ............................ -

Cephalopoda: Loliginidae and Idiosepiidae)
Marine Biology (2005) 147: 1323–1332 DOI 10.1007/s00227-005-0014-5 RESEARCH ARTICLE Delphine Pichon Æ Valeria Gaia Æ Mark D. Norman Renata Boucher-Rodoni Phylogenetic diversity of epibiotic bacteria in the accessory nidamental glands of squids (Cephalopoda: Loliginidae and Idiosepiidae) Received: 25 August 2004 / Accepted: 14 April 2005 / Published online: 30 July 2005 Ó Springer-Verlag 2005 Abstract Bacterial communities were identified from the ginids) are associated with Silicibacter-related strains, accessory nidamental glands (ANGs) of European and suggesting a biogeographic clustering for the Agrobac- Western Pacific squids of the families Loliginidae terium-like strains. and Idiosepiidae, as also in the egg capsules, embryo and yolk of two loliginid squid species, and in the entire egg of one idiosepiid squid species. The results of phyloge- netic analyses of 16S RNA gene (rDNA) confirmed that Introduction several phylotypes of a-proteobacteria, c-proteobacteria and Cytophaga-Flavobacteria-Bacteroides phylum were The female reproductive systems of certain cephalopod present as potential symbiotic associations within the taxa possess a paired organ associated with egg laying ANGs. Several identified clones were related to reference that contains dense bacterial communities (Bloodgood strains, while others had no known close relatives. Gram 1977). Known as the ‘‘accessory nidamental glands’’ positive strains were rare in loliginid squids. Several (ANGs), these organs are present in pencil and reef squids bacterial groups may play important roles in the func- (family Loliginidae), cuttlefishes (family Sepiidae), bob- tion of the ANGs, such as production of the toxic tail squids (family Sepiolidae), bottletail squids (family compounds involved in egg protection and carotenoid Sepiadariidae) and pygmy squids (family Idiosepiidae). -

Scientific Reports 7: 11033
www.nature.com/scientificreports Correction: Author Correction OPEN Introduced ascidians harbor highly diverse and host-specifc symbiotic microbial assemblages Received: 11 May 2017 James S. Evans1, Patrick M. Erwin 1, Noa Shenkar2 & Susanna López-Legentil1 Accepted: 22 August 2017 Many ascidian species have experienced worldwide introductions, exhibiting remarkable success Published: xx xx xxxx in crossing geographic borders and adapting to local environmental conditions. To investigate the potential role of microbial symbionts in these introductions, we examined the microbial communities of three ascidian species common in North Carolina harbors. Replicate samples of the globally introduced species Distaplia bermudensis, Polyandrocarpa anguinea, and P. zorritensis (n = 5), and ambient seawater (n = 4), were collected in Wrightsville Beach, NC. Microbial communities were characterized by next-generation (Illumina) sequencing of partial (V4) 16S rRNA gene sequences. Ascidians hosted diverse symbiont communities, consisting of 5,696 unique microbial OTUs (at 97% sequenced identity) from 44 bacterial and three archaeal phyla. Permutational multivariate analyses of variance revealed clear diferentiation of ascidian symbionts compared to seawater bacterioplankton, and distinct microbial communities inhabiting each ascidian species. 103 universal core OTUs (present in all ascidian replicates) were identifed, including taxa previously described in marine invertebrate microbiomes with possible links to ammonia-oxidization, denitrifcation, pathogenesis, and heavy-metal processing. These results suggest ascidian microbial symbionts exhibit a high degree of host-specifcity, forming intimate associations that may contribute to host adaptation to new environments via expanded tolerance thresholds and enhanced holobiont function. Modern society is founded upon the rapid transgression of people and goods across geographic borders; however, this globalization has come at an ecological cost: the introduction of nonnative species1. -
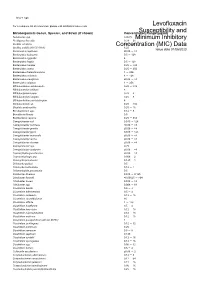
Susceptibility and Resistance Data
toku-e logo For a complete list of references, please visit antibiotics.toku-e.com Levofloxacin Microorganism Genus, Species, and Strain (if shown) Concentration Range (μg/ml)Susceptibility and Aeromonas spp. 0.0625 Minimum Inhibitory Alcaligenes faecalis 0.39 - 25 Bacillus circulans Concentration0.25 - 8 (MIC) Data Bacillus subtilis (ATCC 6051) 6.25 Issue date 01/06/2020 Bacteroides capillosus ≤0.06 - >8 Bacteroides distasonis 0.5 - 128 Bacteroides eggerthii 4 Bacteroides fragilis 0.5 - 128 Bacteroides merdae 0.25 - >32 Bacteroides ovatus 0.25 - 256 Bacteroides thetaiotaomicron 1 - 256 Bacteroides uniformis 4 - 128 Bacteroides ureolyticus ≤0.06 - >8 Bacteroides vulgatus 1 - 256 Bifidobacterium adolescentis 0.25 - >32 Bifidobacterium bifidum 8 Bifidobacterium breve 0.25 - 8 Bifidobacterium longum 0.25 - 8 Bifidobacterium pseudolongum 8 Bifidobacterium sp. 0.25 - >32 Bilophila wadsworthia 0.25 - 16 Brevibacterium spp. 0.12 - 8 Brucella melitensis 0.5 Burkholderia cepacia 0.25 - 512 Campylobacter coli 0.015 - 128 Campylobacter concisus ≤0.06 - >8 Campylobacter gracilis ≤0.06 - >8 Campylobacter jejuni 0.015 - 128 Campylobacter mucosalis ≤0.06 - >8 Campylobacter rectus ≤0.06 - >8 Campylobacter showae ≤0.06 - >8 Campylobacter spp. 0.25 Campylobacter sputorum ≤0.06 - >8 Capnocytophaga ochracea ≤0.06 - >8 Capnocytophaga spp. 0.006 - 2 Chlamydia pneumonia 0.125 - 1 Chlamydia psittaci 0.5 Chlamydia trachomatis 0.12 - 1 Chlamydophila pneumonia 0.5 Citrobacter diversus 0.015 - 0.125 Citrobacter freundii ≤0.00625 - >64 Citrobacter koseri 0.015 - -

Isolation of Lactic-Acid Related Bacteria from the Pig Mucosal P
Aus dem Institut für Tierernährung des Fachbereichs Veterinärmedizin der Freien Universität Berlin Isolation of lactic acid-related bacteria from the pig mucosal proximal gastrointestinal tract, including Olsenella umbonata sp. nov. and Veillonella magna sp. nov. Inaugural-Dissertation zur Erlangung des Grades eines Doktors der Veterinärmedizin an der Freien Universität Berlin vorgelegt von Mareike Kraatz Tierärztin aus Berlin Berlin 2011 Journal-Nr.: 3431 Gedruckt mit Genehmigung des Fachbereichs Veterinärmedizin der Freien Universität Berlin Dekan: Univ.-Prof. Dr. Leo Brunnberg Erster Gutachter: Univ.-Prof. a. D. Dr. Ortwin Simon Zweiter Gutachter: Univ.-Prof. Dr. Lothar H. Wieler Dritter Gutachter: Univ.-Prof. em. Dr. Dr. h. c. Gerhard Reuter Deskriptoren (nach CAB-Thesaurus): anaerobes; Bacteria; catalase; culture media; digestive tract; digestive tract mucosa; food chains; hydrogen peroxide; intestinal microorganisms; isolation; isolation techniques; jejunum; lactic acid; lactic acid bacteria; Lactobacillus; Lactobacillus plantarum subsp. plantarum; microbial ecology; microbial flora; mucins; mucosa; mucus; new species; Olsenella; Olsenella profusa; Olsenella uli; oxygen; pigs; propionic acid; propionic acid bacteria; species composition; stomach; symbiosis; taxonomy; Veillonella; Veillonella ratti Tag der Promotion: 21. Januar 2011 Diese Dissertation ist als Buch (ISBN 978-3-8325-2789-1) über den Buchhandel oder online beim Logos Verlag Berlin (http://www.logos-verlag.de) erhältlich. This thesis is available as a book (ISBN 978-3-8325-2789-1) -

The Role of Territorial Grazers in Coral Reef Trophic Dynamics from Microbes to Apex Predators
ResearchOnline@JCU This file is part of the following reference: Casey, Jordan Marie (2015) The role of territorial grazers in coral reef trophic dynamics from microbes to apex predators. PhD thesis, James Cook University. Access to this file is available from: http://researchonline.jcu.edu.au/41148/ The author has certified to JCU that they have made a reasonable effort to gain permission and acknowledge the owner of any third party copyright material included in this document. If you believe that this is not the case, please contact [email protected] and quote http://researchonline.jcu.edu.au/41148/ The role of territorial grazers in coral reef trophic dynamics from microbes to apex predators Thesis submitted by Jordan Marie Casey April 2015 For the degree of Doctor of Philosophy ARC Centre of Excellence for Coral Reef Studies College of Marine and Environmental Sciences James Cook University ! Acknowledgements First and foremost, I thank my supervisory team, Sean Connolly, J. Howard Choat, and Tracy Ainsworth, for their continuous intellectual support throughout my time at James Cook University. It was invaluable to draw upon the collective insights and constructive criticisms of an ecological modeller, an ichthyologist, and a microbiologist. I am grateful to each of my supervisors for their unique contributions to this PhD thesis. I owe many thanks to all of the individuals that assisted me in the field: Kristen Anderson, Andrew Baird, Shane Blowes, Simon Brandl, Ashley Frisch, Chris Heckathorn, Mia Hoogenboom, Oona Lönnstedt, Chris Mirbach, Chiara Pisapia, Justin Rizzari, and Melanie Trapon. I also thank the directors and staff of Lizard Island Research Station and the Research Vessel James Kirby for efficiently facilitating my research trips. -

HL7 Implementation Guide for CDA Release 2: NHSN Healthcare Associated Infection (HAI) Reports, Release 2 (U.S
CDAR2L3_IG_HAIRPT_R2_D2_2009FEB HL7 Implementation Guide for CDA Release 2: NHSN Healthcare Associated Infection (HAI) Reports, Release 2 (U.S. Realm) Draft Standard for Trial Use February 2009 © 2009 Health Level Seven, Inc. Ann Arbor, MI All rights reserved HL7 Implementation Guide for CDA R2 NHSN HAI Reports Page 1 Draft Standard for Trial Use Release 2 February 2009 © 2009 Health Level Seven, Inc. All rights reserved. Co-Editor: Marla Albitz Contractor to CDC, Lockheed Martin [email protected] Co-Editor/Co-Chair: Liora Alschuler Alschuler Associates, LLC [email protected] Co-Chair: Calvin Beebe Mayo Clinic [email protected] Co-Chair: Keith W. Boone GE Healthcare [email protected] Co-Editor/Co-Chair: Robert H. Dolin, M.D. Semantically Yours, LLC [email protected] Co-Editor Jonathan Edwards CDC [email protected] Co-Editor Pavla Frazier RN, MSN, MBA Contractor to CDC, Frazier Consulting [email protected] Co-Editor: Sundak Ganesan, M.D. SAIC Consultant to CDC/NCPHI [email protected] Co-Editor: Rick Geimer Alschuler Associates, LLC [email protected] Co-Editor: Gay Giannone M.S.N., R.N. Alschuler Associates LLC [email protected] Co-Editor Austin Kreisler Consultant to CDC/NCPHI [email protected] Primary Editor: Kate Hamilton Alschuler Associates, LLC [email protected] Primary Editor: Brett Marquard Alschuler Associates, LLC [email protected] Co-Editor: Daniel Pollock, M.D. CDC [email protected] HL7 Implementation Guide for CDA R2 NHSN HAI Reports Page 2 Draft Standard for Trial Use Release 2 February 2009 © 2009 Health Level Seven, Inc. All rights reserved. Acknowledgments This DSTU was produced and developed in conjunction with the Division of Healthcare Quality Promotion, National Center for Preparedness, Detection, and Control of Infectious Diseases (NCPDCID), and Centers for Disease Control and Prevention (CDC). -

Phylogenetic Analysis of Secretion Systems in Francisellaceae and Legionellales
Phylogenetic analysis of secretion systems in Francisellaceae and Legionellales Investigating events of intracellularization Karl Nyrén Degree project in bioinformatics, 2021 Examensarbete i bioinformatik 45 hp till masterexamen, 2021 Biology Education Centre and IBG, Uppsala University Supervisors: Lionel Guy and Andrei Guliaev Abstract Host-adapted bacteria are pathogens that, through evolutionary time and host-adaptive events, acquired the ability to manipulate hosts into assisting their own reproduction and spread. Through these host-adaptive events, free-living pathogens may be rendered unable to reproduce without their host, which is an irreversible step in evolution. Francisellaceae and Legionellales, two orders of Gammaproteobacteria, are cases where host-adaptation has lead to an intracellular lifestyle. Both orders use secretion systems, in combination with effector proteins, to invade and control their hosts. A current view is that Francisellaceae and Legionellales went through host-adaptive events at two separate time points. However, F. hongkongensis, a member of Francisellaceae shares the same secretion system as the order of Legionellales. Additionally, two host-adapted Gammaproteobacteria, Piscirickettsia spp. and Berkiella spp., swaps phylogenetic position between Legionellales and Francisellaceae depending on methods applied - indicating shared features of Francisellaceae and Legionellales. In this study, we set up a workflow to screen public metagenomic data for candidate host-adaptive bacteria. Using this data, we attempted to assert the phylogenetic position and possibly resolve evolutionary events that occurred in Legionellales, F. hongkongensis, Francisellaceae, Piscirickettsia spp. and Berkiella spp. We successfully acquired 23 candidate host-adapted MAGs by (i) scanning for genes, among reads before assembly, using PhyloMagnet, and (ii) screening for complete secretion systems with MacSyFinder. -

Downloaded from the Virulence Factor of Pathogenic Bacteria Database (VFDB)70
www.nature.com/scientificreports OPEN Investigation of potential pathogenicity of Willaertia magna by investigating the transfer of bacteria pathogenicity genes into its genome Issam Hasni1,2, Nisrine Chelkha1, Emeline Baptiste1, Mouh Rayane Mameri2, Joel Lachuer3,4,5, Fabrice Plasson2, Philippe Colson 1 & Bernard La Scola1* Willaertia magna c2c maky is a thermophilic amoeba closely related to the genus Naegleria. This free-living amoeba has the ability to eliminate Legionella pneumophila, which is an amoeba-resisting bacterium living in an aquatic environment. To prevent the proliferation of L. pneumophila in cooling towers, the use of W. magna as natural biocide has been proposed. To provide a better understanding of the W. magna genome, whole-genome sequencing was performed through the study of virulence factors and lateral gene transfers. This amoeba harbors a genome of 36.5 megabases with 18,519 predicted genes. BLASTp analyses reported protein homology between 136 W. magna sequences and amoeba-resistant microorganisms. Horizontal gene transfers were observed based on the basis of the phylogenetic reconstruction hypothesis. We detected 15 homologs of N. fowleri genes related to virulence, although these latter were also found in the genome of N. gruberi, which is a non-pathogenic amoeba. Furthermore, the cytotoxicity test performed on human cells supports the hypothesis that the strain c2c maky is a non-pathogenic amoeba. This work explores the genomic repertory for the frst draft genome of genus Willaertia and provides genomic data for further comparative studies on virulence of related pathogenic amoeba, N. fowleri. Heterolobosea is a rich class of species of heterotrophs, almost all free-living, containing pathogenic (e.g. -

Prevotella Jejuni Sp. Nov., Isolated from the Small Intestine of a Child with Coeliac Disease
International Journal of Systematic and Evolutionary Microbiology (2013), 63, 4218–4223 DOI 10.1099/ijs.0.052647-0 Prevotella jejuni sp. nov., isolated from the small intestine of a child with coeliac disease Maria E. Hedberg,1 Anne Israelsson,1 Edward R. B. Moore,2,3 Liselott Svensson-Stadler,2 Sun Nyunt Wai,4 Grzegorz Pietz,1 Olof Sandstro¨m,5 Olle Hernell,5 Marie-Louise Hammarstro¨m1 and Sten Hammarstro¨m1 Correspondence 1Department of Clinical Microbiology, Immunology, Umea˚ University, SE-90187 Umea˚, Sweden Maria E. Hedberg 2CCUG – Culture Collection University of Gothenburg, Department of Clinical Bacteriology, [email protected] Sahlgrenska University Hospital, SE-41345 Go¨teborg, Sweden Sten Hammarstro¨m 3 [email protected] Department of Infectious Diseases, Sahlgrenska Academy of the University of Gothenburg, SE-40530 Go¨teborg, Sweden 4Department of Molecular Biology, Umea˚ University, SE-90187 Umea˚, Sweden 5Department of Clinical Sciences, Pediatrics, Umea˚ University, SE-90187 Umea˚, Sweden Five obligately anaerobic, Gram-stain-negative, saccharolytic and proteolytic, non-spore-forming bacilli (strains CD3 : 27, CD3 : 28T, CD3 : 33, CD3 : 32 and CD3 : 34) are described. All five strains were isolated from the small intestine of a female child with coeliac disease. Cells of the five strains were short rods or coccoid cells with longer filamentous forms seen sporadically. The organisms produced acetic acid and succinic acid as major metabolic end products. Phylogenetic analysis based on comparative 16S rRNA gene sequence analysis revealed close relationships between CD3 : 27, CD3 : 28T and CD3 : 33, between CD3 : 32 and Prevotella histicola CCUG 55407T, and between CD3 : 34 and Prevotella melaninogenica CCUG 4944BT.