Encapsidated Hepatitis B Virus Reverse Transcriptase Is Poised on an Ordered RNA Lattice
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Immunogenicity and Efficacy Testing in Chimpanzees of an Oral Hepatitis B Vaccine Based on Live Recombinant Adenovirus
Proc. Natl. Acad. Sci. USA Vol. 86, pp. 6763-6767, September 1989 Medical Sciences Immunogenicity and efficacy testing in chimpanzees of an oral hepatitis B vaccine based on live recombinant adenovirus (adenovirus animal model/adenovirus-vectored vacdnes) MICHAEL D. LUBECK*, ALAN R. DAVIS*, MURTY CHENGALVALA*, ROBERT J. NATUK*, JOHN E. MORIN*, KATHERINE MOLNAR-KIMBER*, BRUCE B. MASON*, BHEEM M. BHAT*, SATOSHI MIZUTANI*, PAUL P. HUNG*, AND ROBERT H. PURCELLO *Wyeth-Ayerst Research, Biotechnology and Microbiology Division, P.O. Pox 8299, Philadelphia, PA 19101; and tLaboratory of Infectious Diseases, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD 20892 Contributed by Robert H. Purcell, May 30, 1989 ABSTRACT As a major cause of acute and chronic liver which, because oftheir wide prior usage, are good candidates disease as well as hepatocellular carcinoma, hepatitis B virus as vectors. Other less well-characterized small animal models (HBV) continues to pose significant health problems world- for human adenoviruses have been occasionally reported wide. Recombinant hepatitis B vaccines based on adenovirus (11-13) but are likewise nonpermissive for Ad4 and Ad7 vectors have been developed to address global needs for infections (unpublished data). An early study of animal effective control of hepatitits B infection. Although consider- species including nonhuman primates indicated that human able progress has been made in the construction ofrecombinant adenoviruses do not induce acute respiratory disease in adenoviruses that express large amounts of HBV gene prod- monkeys or chimpanzees following intranasal inoculations ucts, preclinical immunogenicity and efficacy testing of candi- (14). Serological data, however, indicated that such experi- date vaccines has remained difficult due to the lack ofa suitable mental infections occasionally induced anti-adenovirus anti- animal model. -

Inhibition of Hepadnaviral Replication by Polyethylenimine-Based Intravenous Delivery of Antisense Phosphodiester Oligodeoxynucleotides to the Liver
Gene Therapy (2001) 8, 874–881 2001 Nature Publishing Group All rights reserved 0969-7128/01 $15.00 www.nature.com/gt RESEARCH ARTICLE Inhibition of hepadnaviral replication by polyethylenimine-based intravenous delivery of antisense phosphodiester oligodeoxynucleotides to the liver M Robaczewska1,2, S Guerret3, J-S Remy4, I Chemin1, W-B Offensperger5, M Chevallier6, J-P Behr4, AJ Podhajska2, HE Blum5, C Trepo1 and L Cova1 1INSERM U271, Lyon, France; 2Faculty of Biotechnology, University of Gdansk and Medical University of Gdansk, Gdansk, Poland; 3Biomaterials Laboratory, Faculty of Pharmacy, Lyon; 4Faculty of Pharmacy, Illkirch, France; 5Department of Medicine, University of Freiburg, Germany; and 6Laboratoires Marcel Me´rieux, Lyon, France Antisense oligodeoxynucleotides (ODNs) appear as attract- fold as compared with the O-ODN-AS2. Following 9-day ive anti-hepatitis B virus (HBV) agents. We investigated in therapy the intrahepatic levels of both DHBV DNA and RNA vivo, in the duck HBV (DHBV) infection model, whether lin- were significantly decreased in the lPEI/O-ODN-AS2-treated ear polyethylenimine (lPEI)-based intravenous delivery of group as compared with the O-ODN-AS2-treated, control the natural antisense phosphodiester ODNs (O-ODNs) can lPEI/O-ODN-treated, and untreated controls. In addition, prevent their degradation and allow viral replication inhibition inhibition of intrahepatic viral replication by lPEI/O-ODN-AS2 in the liver. DHBV-infected Pekin ducklings were injected was not associated with toxicity and was comparable with with antisense O-ODNs covering the initiation codon of the that induced by the phosphorothioate S-ODN-AS2 at a five- DHBV large envelope protein, either in free form (O-ODN- fold higher dose. -

Hepatitis B Fast Facts Everything You Need to Know in 2 Minutes Or Less!
Hepatitis B Foundation Cause for a Cure www.hepb.org Hepatitis B Fast Facts Everything you need to know in 2 minutes or less! Hepatitis B is the most common serious liver infection in the world. It is caused by the hepatitis B virus (HBV) that attacks liver cells and can lead to liver failure, cirrhosis (scarring) or cancer of the liver. The virus is transmitted through contact with blood and bodily fluids that contain blood. Most people are able to fight off the hepatitis B infection and clear the virus from their blood. This may take up to six months. While the virus is present in their blood, infected people can pass the virus on to others. Approximately 5-10% of adults, 30-50% of children, and 90% of babies will not get rid of the virus and will develop chronic infection. Chronically infected people can pass the virus on to others and are at increased risk for liver problems later in life. The hepatitis B virus is 100 times more infectious than the AIDS virus. Yet, hepatitis B can be pre- vented with a safe and effective vaccine. For the 400 million people worldwide who are chronically infected with hepatitis B, the vaccine is of no use. However, there are promising new treatments for those who live with chronic hepatitis B. In the World: • This year alone, 10 to 30 million people will become infected with the hepatitis B virus (HBV). • The World Health Organization estimates that 400 million people worldwide are already chronically infected with hepatitis B. -

Prevention & Control of Viral Hepatitis Infection
Prevention & Control of Viral Hepatitis Infection: A Strategy for Global Action © World Health Organization 2011. All rights reserved. The designations employed and the presentation of the material in this publication do not imply the expression of any opinion whatsoever on the part of the World Health Organization concerning the legal status of any country, territory, city or area or of its authorities, or concerning the delimitation of its frontiers or boundaries. Dotted lines on maps represent approximate border lines for which there may not yet be full agreement. The mention of specific companies or of certain manufacturers’ products does not imply that they are endorsed or recommended by the World Health Organization in preference to others of a similar nature that are not mentioned. Errors and omissions excepted, the names of proprietary products are distinguished by initial capital letters. All reasonable precautions have been taken by WHO to verify the information contained in this publication. However, the published material is being distributed without warranty of any kind, either express or implied. The responsibility for the interpretation and use of the material lies with the reader. In no event shall the World Health Organization be liable for damages arising from its use. Table of contents Disease burden 02 What is viral hepatitis? 05 Prevention & control: a tailored approach 06 Global Achievements 08 Remaining challenges 10 World Health Assembly: a mandate for comprehensive prevention & control 13 WHO goals and strategy -

HBV Cccdna: Viral Persistence Reservoir and Key Obstacle for a Cure of Chronic Hepatitis B Michael Nassal
Downloaded from http://gut.bmj.com/ on June 8, 2015 - Published by group.bmj.com Gut Online First, published on June 5, 2015 as 10.1136/gutjnl-2015-309809 Recent advances in basic science HBV cccDNA: viral persistence reservoir and key obstacle for a cure of chronic hepatitis B Michael Nassal Correspondence to ABSTRACT frequent viral rebound upon therapy withdrawal Dr Michael Nassal, Department At least 250 million people worldwide are chronically indicates a need for lifelong treatment.6 of Internal Medicine II/ Molecular Biology, University infected with HBV, a small hepatotropic DNA virus that Reactivation can even occur, upon immunosuppres- Hospital Freiburg, Hugstetter replicates through reverse transcription. Chronic infection sion, in patients who resolved an acute HBV infec- Str. 55, Freiburg D-79106, greatly increases the risk for terminal liver disease. tion decades ago,7 indicating that the virus can be Germany; Current therapies rarely achieve a cure due to the immunologically controlled but is not eliminated. [email protected] refractory nature of an intracellular viral replication The virological key to this persistence is an intra- Received 17 April 2015 intermediate termed covalently closed circular (ccc) DNA. cellular HBV replication intermediate, called cova- Revised 12 May 2015 Upon infection, cccDNA is generated as a plasmid-like lently closed circular (ccc) DNA, which resides in Accepted 13 May 2015 episome in the host cell nucleus from the protein-linked the nucleus of infected cells as an episomal (ie, relaxed circular (RC) DNA genome in incoming virions. non-integrated) plasmid-like molecule that gives Its fundamental role is that as template for all viral rise to progeny virus. -

Hepatitis B? HEPATITIS B Hepatitis B Is a Contagious Liver Disease That Results from Infection with the Hepatitis B Virus
What is Hepatitis B? HEPATITIS B Hepatitis B is a contagious liver disease that results from infection with the Hepatitis B virus. When first infected, a person can develop Are you at risk? an “acute” infection, which can range in severity from a very mild illness with few or no symptoms to a serious condition requiring hospitalization. Acute Hepatitis B refers to the first 6 months after someone is exposed to the Hepatitis B virus. Some people are able to fight the infection and clear the virus. For others, the infection remains and leads to a “chronic,” or lifelong, illness. Chronic Hepatitis B refers to the illness that occurs when the Hepatitis B virus remains in a person’s body. Over time, the infection can cause serious health problems. How is Hepatitis B spread? Hepatitis B is usually spread when blood, semen, or other body fluids from a person infected with the Hepatitis B virus enter the body of someone who is not infected. This can happen through having sex with an infected partner; sharing needles, syringes, or other injection drug equipment; or from direct contact with the blood or open sores of an infected person. Hepatitis B can also be passed from an infected mother to her baby at birth. Who should be tested for Hepatitis B? Approximately 1.2 million people in the United States and 350 million people worldwide have Hepatitis B. Testing for Hepatitis B is recommended for certain groups of people, including: Most are unaware of their infection. ■ People born in Asia, Africa, and other regions with moderate or high rates Is Hepatitis B common? of Hepatitis B (see map) Yes. -
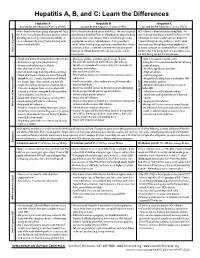
Hepatitis A, B, and C: Learn the Differences
Hepatitis A, B, and C: Learn the Differences Hepatitis A Hepatitis B Hepatitis C caused by the hepatitis A virus (HAV) caused by the hepatitis B virus (HBV) caused by the hepatitis C virus (HCV) HAV is found in the feces (poop) of people with hepa- HBV is found in blood and certain body fluids. The virus is spread HCV is found in blood and certain body fluids. The titis A and is usually spread by close personal contact when blood or body fluid from an infected person enters the body virus is spread when blood or body fluid from an HCV- (including sex or living in the same household). It of a person who is not immune. HBV is spread through having infected person enters another person’s body. HCV can also be spread by eating food or drinking water unprotected sex with an infected person, sharing needles or is spread through sharing needles or “works” when contaminated with HAV. “works” when shooting drugs, exposure to needlesticks or sharps shooting drugs, through exposure to needlesticks on the job, or from an infected mother to her baby during birth. or sharps on the job, or sometimes from an infected How is it spread? Exposure to infected blood in ANY situation can be a risk for mother to her baby during birth. It is possible to trans- transmission. mit HCV during sex, but it is not common. • People who wish to be protected from HAV infection • All infants, children, and teens ages 0 through 18 years There is no vaccine to prevent HCV. -

Understanding Human Astrovirus from Pathogenesis to Treatment
University of Tennessee Health Science Center UTHSC Digital Commons Theses and Dissertations (ETD) College of Graduate Health Sciences 6-2020 Understanding Human Astrovirus from Pathogenesis to Treatment Virginia Hargest University of Tennessee Health Science Center Follow this and additional works at: https://dc.uthsc.edu/dissertations Part of the Diseases Commons, Medical Sciences Commons, and the Viruses Commons Recommended Citation Hargest, Virginia (0000-0003-3883-1232), "Understanding Human Astrovirus from Pathogenesis to Treatment" (2020). Theses and Dissertations (ETD). Paper 523. http://dx.doi.org/10.21007/ etd.cghs.2020.0507. This Dissertation is brought to you for free and open access by the College of Graduate Health Sciences at UTHSC Digital Commons. It has been accepted for inclusion in Theses and Dissertations (ETD) by an authorized administrator of UTHSC Digital Commons. For more information, please contact [email protected]. Understanding Human Astrovirus from Pathogenesis to Treatment Abstract While human astroviruses (HAstV) were discovered nearly 45 years ago, these small positive-sense RNA viruses remain critically understudied. These studies provide fundamental new research on astrovirus pathogenesis and disruption of the gut epithelium by induction of epithelial-mesenchymal transition (EMT) following astrovirus infection. Here we characterize HAstV-induced EMT as an upregulation of SNAI1 and VIM with a down regulation of CDH1 and OCLN, loss of cell-cell junctions most notably at 18 hours post-infection (hpi), and loss of cellular polarity by 24 hpi. While active transforming growth factor- (TGF-) increases during HAstV infection, inhibition of TGF- signaling does not hinder EMT induction. However, HAstV-induced EMT does require active viral replication. -

Hepatitis B Factsheet: COVID-19 and Hep B
Hepatitis B factsheet: COVID-19 and hep B For more information about anything in this factsheet, phone the Hepatitis Infoline on 1800 803 990 or go to www.hep.org.au COVID-19 and hep B Frequently Asked Questions (FAQ) There’s so much information swirling around these days as we deal with the changing world around us due to COVID-19. There’s lots of incorrect information and it’s easy to be overwhelmed and confused. On this page, we’ll try to clear some things up. It is important that information is helpful, clear, and checked and confirmed by experts. Please share the following information – it comes from health and medical specialists. COVID-19 is an acronym that stands for COronaVIrus Disease 2019. You might hear it called coronavirus, corona, SARS-CoV-2, or even rona. We’ll call it COVID-19 or coronavirus in this FAQ but all these terms are essentially talking about the same thing and we’ll explain them at the bottom. You’ll also hear new or unfamiliar terms like “social distancing,” “physical distancing,” “self-isolation,” and “quarantine”. These terms and what they actually mean can become confusing so we’ve including a glossary at the bottom of this FAQ. This virus has only been known about since around December 2019 so it’s all very new, information changes quickly, and we’re all learning day-by-day and week-by-week. Compare COVID-19 to hep B which we’ve known about since the 1980s and hep B even earlier in the 1960s! We’ll regularly update this page as new information comes to light. -

Interaction Between Hepatitis B Virus and SARS-Cov-2 Infections
World Journal of W J G Gastroenterology Submit a Manuscript: https://www.f6publishing.com World J Gastroenterol 2021 March 7; 27(9): 782-793 DOI: 10.3748/wjg.v27.i9.782 ISSN 1007-9327 (print) ISSN 2219-2840 (online) MINIREVIEWS Interaction between hepatitis B virus and SARS-CoV-2 infections Tian-Dan Xiang, Xin Zheng ORCID number: Tian-Dan Xiang Tian-Dan Xiang, Xin Zheng, Department of Infectious Diseases, Union Hospital, Tongji Medical 0000-0003-2792-6631; Xin Zheng College, Huazhong University of Science and Technology, Wuhan 430022, Hubei Province, 0000-0001-6564-7807. China Author contributions: Zheng X Corresponding author: Xin Zheng, MD, PhD, Professor, Department of Infectious Diseases, contributed to the conception and Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, No. design of the work and revised the 1277 Jiefang Avenue, Wuhan 430022, Hubei Province, China. [email protected] manuscript; Xiang TD performed the literature review and drafted the manuscript. Abstract Conflict-of-interest statement: Coronavirus disease 2019 (COVID-19) has become a global pandemic and Authors declare no conflict of garnered international attention. The causative pathogen of COVID-19 is severe interest for this article. acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a novel, highly contagious coronavirus. Numerous studies have reported that liver injury is quite Open-Access: This article is an common in patients with COVID-19. Hepatitis B has a worldwide distribution as open-access article that was well as in China. At present, hepatitis B virus (HBV) remains a leading cause of selected by an in-house editor and cirrhosis, liver failure, and hepatocellular carcinoma. -

Hepatitis B Virus (HBV)
HEPATITIS B AND COLLEGE STUDENTS Hepatitis B is a liver disease that results from infection with the Hepatitis B virus (HBV). It can range in severity from a mild illness lasting a few weeks to a serious, lifelong illness. Hepatitis B is usually spread when blood, semen, or another body fluid from a person infected with the Hepatitis B virus enters the body of someone who is not infected. This can happen through sexual contact with an infected person or sharing needles, syringes, or other drug-injection equipment. Hepatitis B can also be passed from an infected mother to her baby at birth. Hepatitis B can be either acute or chronic. Acute HBV infection is a short-term illness that occurs within the first 6 months after someone is exposed to the Hepatitis B virus. Acute infection can — but does not always — lead to chronic infection. Chronic HBV infection is a long-term illness that occurs when the Hepatitis B virus remains in a person’s body. Chronic HBV is a serious disease that can result in long-term health problems, and even death. The best way to prevent HBV infection is by getting vaccinated. How common is hepatitis B in the United States? About 800,000 to 1.4 million persons in the United States have chronic HBV infection. Each year 38,000 more people, mostly young adults, get infected with HBV and almost 2,000 people die from chronic HBV. How is hepatitis B spread? Hepatitis B is spread when blood, semen, or other body fluid infected with the hepatitis B virus enters the body of a person who is not infected. -

Interferon Gene Transfer by a Hepatitis B Virus Vector Efficiently Suppresses Wild-Type Virus Infection
Proc. Natl. Acad. Sci. USA Vol. 96, pp. 10818–10823, September 1999 Medical Sciences Interferon gene transfer by a hepatitis B virus vector efficiently suppresses wild-type virus infection ULRIKE PROTZER*, MICHAEL NASSAL*†,PEI-WEN CHIANG*‡,MICHAEL KIRSCHFINK§, AND HEINZ SCHALLER*¶ *Zentrum fu¨r Molekulare Biologie Heidelberg and §Department of Immunology, University of Heidelberg, Im Neuenheimer Feld, D-69120 Heidelberg, Germany; and †University Hospital, Department of Internal Medicine II͞Molecular Biology, University of Freiburg, Hugstetter Strasse 55, D-79106 Freiburg, Germany Communicated by Peter H. Duesberg, University of California, Berkeley, CA, July 13, 1999 (received for review February 25, 1999) ABSTRACT Hepatitis B viruses specifically target the concepts include gene therapy and the use of defective or liver, where they efficiently infect quiescent hepatocytes. Here attenuated viruses to block wild-type viral infection (3). Im- we show that human and avian hepatitis B viruses can be munomodulatory cytokines such as IL-12, IFN-␥ or tumor converted into vectors for liver-directed gene transfer. These necrosis factor-␣ potently suppress hepatitis B virus (HBV) vectors allow hepatocyte-specific expression of a green fluo- replication in an HBV transgenic mouse model (10, 11), rescent protein in vitro and in vivo. Moreover, when used to whereas IL-12 and the Th1 cytokines IFN-␥ and IL-2 seem to transduce a type I interferon gene, expression of interferon play an important role for viral clearance in chronically efficiently suppresses wild-type virus replication in the duck infected patients (12). However, systemic application of cyto- model of hepatitis B virus infection. These data suggest local kines is limited by severe side effects (7, 9).