Transcriptomic Profile and Defense Strategies in the Red Seaweed Laurencia Dendroidea J
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Rehmannia Glutinosa-Monocultured Rhizosphere Soil
Comparative Metaproteomic Analysis on Consecutively Rehmannia glutinosa-Monocultured Rhizosphere Soil Linkun Wu1,2, Haibin Wang1,2., Zhixing Zhang1,2., Rui Lin2,3, Zhongyi Zhang1,4, Wenxiong Lin1,2* 1 School of Life Sciences, Fujian Agriculture and Forestry University, Fuzhou, Fujian, China, 2 Agroecological Institute, Fujian Agriculture and Forestry University, Fuzhou, Fujian, China, 3 College of Oceanography and Environmental Science, Xiamen University, Xiamen, Fujian, China, 4 Institute of Chinese Medicinal Materials, Henan Agriculture University, Zhengzhou, Henan, China Abstract Background: The consecutive monoculture for most of medicinal plants, such as Rehmannia glutinosa, results in a significant reduction in the yield and quality. There is an urgent need to study for the sustainable development of Chinese herbaceous medicine. Methodology/Principal Findings: Comparative metaproteomics of rhizosphere soil was developed and used to analyze the underlying mechanism of the consecutive monoculture problems of R. glutinosa. The 2D-gel patterns of protein spots for the soil samples showed a strong matrix dependency. Among the spots, 103 spots with high resolution and repeatability were randomly selected and successfully identified by MALDI TOF-TOF MS for a rhizosphere soil metaproteomic profile analysis. These proteins originating from plants and microorganisms play important roles in nutrient cycles and energy flow in rhizospheric soil ecosystem. They function in protein, nucleotide and secondary metabolisms, signal transduction and resistance. Comparative metaproteomics analysis revealed 33 differentially expressed protein spots in rhizosphere soil in response to increasing years of monoculture. Among them, plant proteins related to carbon and nitrogen metabolism and stress response, were mostly up-regulated except a down-regulated protein (glutathione S-transferase) involving detoxification. -
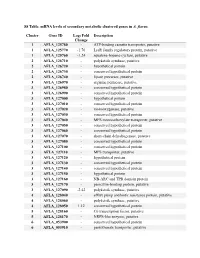
S8 Table. Mrna Levels of Secondary Metabolic Clustered Genes in A
S8 Table. mRNA levels of secondary metabolic clustered genes in A. flavus. Cluster Gene ID Log2 Fold Description Change 1 AFLA_125780 - ATP-binding cassette transporter, putative 1 AFLA_125770 -1.76 LysR family regulatory protein, putative 1 AFLA_125760 -1.24 squalene-hopene-cyclase, putative 2 AFLA_126710 - polyketide synthase, putative 2 AFLA_126720 - hypothetical protein 2 AFLA_126730 - conserved hypothetical protein 2 AFLA_126740 - lipase precursor, putative 3 AFLA_126970 - arginine permease, putative 3 AFLA_126980 - conserved hypothetical protein 3 AFLA_126990 - conserved hypothetical protein 3 AFLA_127000 - hypothetical protein 3 AFLA_127010 - conserved hypothetical protein 3 AFLA_127020 - monooxygenase, putative 3 AFLA_127030 - conserved hypothetical protein 3 AFLA_127040 - MFS monocarboxylate transporter, putative 3 AFLA_127050 - conserved hypothetical protein 3 AFLA_127060 - conserved hypothetical protein 3 AFLA_127070 - short-chain dehydrogenase, putative 3 AFLA_127080 - conserved hypothetical protein 3 AFLA_127100 - conserved hypothetical protein 3 AFLA_127110 - MFS transporter, putative 3 AFLA_127120 - hypothetical protein 3 AFLA_127130 - conserved hypothetical protein 3 AFLA_127140 - conserved hypothetical protein 3 AFLA_127150 - hypothetical protein 3 AFLA_127160 - NB-ARC and TPR domain protein 3 AFLA_127170 - penicillin-binding protein, putative 3 AFLA_127090 -2.42 polyketide synthase, putative 4 AFLA_128040 - efflux pump antibiotic resistance protein, putative 4 AFLA_128060 - polyketide synthase, putative 4 AFLA_128050 -

The Life Cycle and Genetic Structure of the Red Alga Furcellaria Lumbricalis on a Salinity Gradient
WALTER AND ANDRÉE DE NOTTBECK FOUNDATION SCIENTIFIC REPORTS View metadata, citation and similar papers at core.ac.uk brought to you by CORE No. 33 provided by Helsingin yliopiston digitaalinen arkisto The life cycle and genetic structure of the red alga Furcellaria lumbricalis on a salinity gradient KIRSI KOSTAMO Academic dissertation in Ecology, to be presented, with the permission of the Faculty of Biosciences of the University of Helsinki, for public criticism in the Auditorium 1041, Biocenter 2, University of Helsinki, Viikinkaari 5, Helsinki, on January 25th, 2008, at 12 noon. HELSINKI 2008 This thesis is based on the following papers, which will be referred to in the text by their Roman numerals: I. Kostamo, K. & Mäkinen, A. 2006: Observations on the mode and seasonality of reproduction in Furcellaria lumbricalis (Gigartinales, Rhodophyta) populations in the northern Baltic Sea. – Bot. Mar. 49: 304-309. II Korpelainen, H., Kostamo, K. & Virtanen, V. 2007: Microsatellite marker identifi cation using genome screening and restriction-ligation. – BioTechniques 42: 479-486. III Kostamo, K., Korpelainen, H., Maggs, C. A. & Provan, J. 2007: Genetic variation among populations of the red alga Furcellaria lumbricalis in northern Europe. – Manuscript. IV Kostamo, K. & Korpelainen, H. 2007: Clonality and small-scale genetic diversity within populations of the red alga Furcellaria lumbricalis (Rhodophyta) in Ireland and in the northern Baltic Sea. – Manuscript. Paper I is printed with permission from Walter de Gruyter Publishers and paper II with permission from BioTechniques. Reviewers: Dr. Elina Leskinen Department of Biological and Environmental Sciences University of Helsinki Finland Prof. Kerstin Johannesson Tjärnö Marine Biological Laboratory University of Gothenburg Sweden Opponent: Dr. -

Chemical Composition and Potential Practical Application of 15 Red Algal Species from the White Sea Coast (The Arctic Ocean)
molecules Article Chemical Composition and Potential Practical Application of 15 Red Algal Species from the White Sea Coast (the Arctic Ocean) Nikolay Yanshin 1, Aleksandra Kushnareva 2, Valeriia Lemesheva 1, Claudia Birkemeyer 3 and Elena Tarakhovskaya 1,4,* 1 Department of Plant Physiology and Biochemistry, Faculty of Biology, St. Petersburg State University, 199034 St. Petersburg, Russia; [email protected] (N.Y.); [email protected] (V.L.) 2 N. I. Vavilov Research Institute of Plant Industry, 190000 St. Petersburg, Russia; [email protected] 3 Faculty of Chemistry and Mineralogy, University of Leipzig, 04103 Leipzig, Germany; [email protected] 4 Vavilov Institute of General Genetics RAS, St. Petersburg Branch, 199034 St. Petersburg, Russia * Correspondence: [email protected] Abstract: Though numerous valuable compounds from red algae already experience high demand in medicine, nutrition, and different branches of industry, these organisms are still recognized as an underexploited resource. This study provides a comprehensive characterization of the chemical composition of 15 Arctic red algal species from the perspective of their practical relevance in medicine and the food industry. We show that several virtually unstudied species may be regarded as promis- ing sources of different valuable metabolites and minerals. Thus, several filamentous ceramialean algae (Ceramium virgatum, Polysiphonia stricta, Savoiea arctica) had total protein content of 20–32% of dry weight, which is comparable to or higher than that of already commercially exploited species Citation: Yanshin, N.; Kushnareva, (Palmaria palmata, Porphyra sp.). Moreover, ceramialean algae contained high amounts of pigments, A.; Lemesheva, V.; Birkemeyer, C.; macronutrients, and ascorbic acid. Euthora cristata (Gigartinales) accumulated free essential amino Tarakhovskaya, E. -

Download PDF Version
MarLIN Marine Information Network Information on the species and habitats around the coasts and sea of the British Isles A red seaweed (Furcellaria lumbricalis) MarLIN – Marine Life Information Network Biology and Sensitivity Key Information Review Will Rayment 2008-05-22 A report from: The Marine Life Information Network, Marine Biological Association of the United Kingdom. Please note. This MarESA report is a dated version of the online review. Please refer to the website for the most up-to-date version [https://www.marlin.ac.uk/species/detail/1616]. All terms and the MarESA methodology are outlined on the website (https://www.marlin.ac.uk) This review can be cited as: Rayment, W.J. 2008. Furcellaria lumbricalis A red seaweed. In Tyler-Walters H. and Hiscock K. (eds) Marine Life Information Network: Biology and Sensitivity Key Information Reviews, [on-line]. Plymouth: Marine Biological Association of the United Kingdom. DOI https://dx.doi.org/10.17031/marlinsp.1616.2 The information (TEXT ONLY) provided by the Marine Life Information Network (MarLIN) is licensed under a Creative Commons Attribution-Non-Commercial-Share Alike 2.0 UK: England & Wales License. Note that images and other media featured on this page are each governed by their own terms and conditions and they may or may not be available for reuse. Permissions beyond the scope of this license are available here. Based on a work at www.marlin.ac.uk (page left blank) Date: 2008-05-22 A red seaweed (Furcellaria lumbricalis) - Marine Life Information Network See online review for distribution map The seaweed Furcellaria lumbricalis, plant with fertile branches. -

The Genus Laurencia (Rhodomelaceae, Rhodophyta) in the Canary Islands
Courier Forsch.-Inst. Senckenberg, 159: 113-117 Frankfurt a.M., 01.07.1993 The Genus Laurencia (Rhodomelaceae, Rhodophyta) in the Canary Islands M. CANDELARIA GIL-RODRIGUEZ & RICARDO HAROUN I Figure The genus Laurencia LAMOUROUX is a group of In the Atlantic Coasts, SAITO (1982) made a short medium-sized, erect, fleshy or cartilaginous, red al review of three typical European species: L. obtusa gae distributed from temperate to tropical waters. (HUDSON) LAMOUROUX, L. pinnatifida (HUDSON) LA During the last few years several collections along MOUROUX and L. hybrida (De.) LENORMAND. Other the coasts of the Canary Islands have shown the im researches carried out in this troublesome genus in portant role of the Laurencia species in the intertidal the Atlantic Ocean were made by TAYLOR (1960) in communities; however, it is rather problematic to Eastern Tropical and Subtropical Coasts of America, identify many of the taxa observed, and it seems 0LIVEIRA-FILHO {1969) in Brazil, MAGNE (1980) in necessary to make a biosystematic review of this ge the French Atlantic Coasts, LAWSON & JoHN (1982) nus in the Macaronesian Region. in the West Coast of Africa, RODRIGUEZ DE Rros LAMouRoux in 1813 established the genus with 8 (1981), RODRIGUEZ DE RIOS & SAITO (1982, 1985) and species, but he didn't mention a type species. Critical RODRIGUEZ DE RIOS & LOBO {1984) in Venezuela. systematic studies have been made by several authors, C. AGARDH (1823, 1824), J.AGARDH {1842, 1851, 1880), DE TONI {1903, 1924), YAMADA {1931), after reviewing many type specimens -

On the Regiospecificity of Vanadium Bromoperoxidase
J. Am. Chem. Soc. 2001, 123, 3289-3294 3289 On the Regiospecificity of Vanadium Bromoperoxidase Jennifer S. Martinez, Georgia L. Carroll, Richard A. Tschirret-Guth, Gereon Altenhoff, R. Daniel Little, and Alison Butler* Contribution from the Department of Chemistry and Biochemistry, UniVersity of California, Santa Barbara, California 93106-9510 ReceiVed December 5, 2000 Abstract: Vanadium haloperoxidase enzymes catalyze the oxidation of halide ions by hydrogen peroxide, producing an oxidized intermediate, which can halogenate an organic substrate or react with a second equivalent of hydrogen peroxide to produce dioxygen. Haloperoxidases are thought to be involved in the biogenesis of halogenated natural products isolated from marine organisms, including indoles and terpenes, of which many are selectively oxidized or halogenated. Little has been shown concerning the ability of the marine haloperoxidases to catalyze regioselective reactions. Here we report the regiospecific bromoperoxidative oxidation of 1,3-di-tert-butylindole by V-BrPO from the marine algae Ascophyllum nodosum and Corallina officinalis. Both enzymes catalyze the regiospecific oxidation of 1,3-di-tert-butylindole in a reaction requiring - both H2O2 and Br as substrates, but which produce the unbrominated 1,3-di-tert-butyl-2-indolinone product exclusively, in near quantitative yield (i.e. one H2O2 consumed per product). By contrast, reactions with the - controlled addition of aqueous bromine solution (HOBr ) Br2 ) Br3 ) produce three monobromo and one dibromo-2-indolinone products, all of which differ from the V-BrPO-catalyzed product. Further, reactivities of 1,3-di-tert-butyl-2-indolinone with both aqueous bromine and V-BrPO differ significantly and shed light onto the possible nature of the oxidizing intermediate. -

Is the Red Alga Meristotheca Papulosa Annual? -Monitoring Of
Aquacult. Sci. 67(1),49-56(2019) Is the red alga Meristotheca papulosa annual? - Monitoring of tagged thalli at Banda, Tateyama, Central Pacific coast of Japan- 1,* 1, 2 1 1 Boryuan CHEN , Shingo AKITA , Akito UEHARA and Daisuke FUJITA Abstract: Meristotheca papulosa is a commercially important red alga used for human consumption. It has been reported as an annual species in Kagoshima Prefecture but has been suggested to be perennial in Kochi Prefecture. In the present study, thalli were indirectly monitored at Banda, Chiba Prefecture. A caged culture and feeding test recording were also conducted. Among the nine thalli tagged in March or April 2016, three survived the winter but disappeared in July 2017. After reaching a maximum length in May, the thallus size decreased from June to November 2016. However, the thalli began to regrow in December 2016. Bite marks were common on the thalli; appearance of herbivorous fishes was recorded by the interval camera. An in-situ culture was conducted by transplanting six thalli each inside and outside of a cage from March to October 2016. After reaching the maximum weight in June, thalli located outside of the cage disappeared in July, but those located inside of the cage survived until September. Aplysia parvula, which was able to intrude the cage, were noted to be influential grazers on the thalli. These results suggest that M. papulosa is perennial but its longevity is affected by wave action and grazing. Key words: Meristotheca papulosa; Perennial; Caging; Grazer were Kagoshima Prefecture in Kyushu (250 to Introduction 300 tons/year) and Izu Islands on the central Pacific coast of Japan (200 to 300 tons/year) Meristotheca papulosa (Montagne) J. -

SEAWEED in the TROPICAL SEASCAPE Stina Tano
SEAWEED IN THE TROPICAL SEASCAPE Stina Tano Seaweed in the tropical seascape Importance, problems and potential Stina Tano ©Stina Tano, Stockholm University 2016 Cover photo: Eucheuma denticulatum and Ulva sp. All photos in the thesis by the author. ISBN 978-91-7649-396-0 Printed in Sweden by Holmbergs, Malmö 2016 Distributor: Department of Ecology, Environment and Plant Science To Johan I may not have gone where I intended to go, but I think I have ended up where I intended to be. Douglas Adams ABSTRACT The increasing demand for seaweed extracts has led to the introduction of non-native seaweeds for farming purposes in many tropical regions. Such intentional introductions can lead to spread of non-native seaweeds from farming areas, which can become established in and alter the dynamics of the recipient ecosystems. While tropical seaweeds are of great interest for aquaculture, and have received much attention as pests in the coral reef literature, little is known about the problems and potential of natural populations, or the role of natural seaweed beds in the tropical seascape. This thesis aims to investigate the spread of non-native genetic strains of the tropical macroalga Eucheuma denticulatum, which have been intentionally introduced for seaweed farming purposes in East Africa, and to evaluate the state of the genetically distinct but morphologically similar native populations. Additionally it aims to investigate the ecological role of seaweed beds in terms of the habitat utilization by fish and mobile invertebrate epifauna. The thesis also aims to evaluate the potential of native populations of eucheumoid seaweeds in regard to seaweed farming. -

Electric Supporting Information (ESI) Crystal Structure and Functional
Electronic Supplementary Material (ESI) for Chemical Science. This journal is © The Royal Society of Chemistry 2018 Electric Supporting Information (ESI) Crystal structure and functional analysis of large-terpene synthase belonging to a newly found subclass Masahiro Fujihashi,a* Tsutomu Sato,b* Yuma Tanaka,a Daisuke Yamamoto,a Tomoyuki Nishi,b Daijiro Ueda,b Mizuki Murakami,b Yoko Yasuno,c Ai Sekihara,c Kazuma Fuku,c Tetsuro Shinadac & Kunio Mikia* a. Department of Chemistry, Graduate School of Science, Kyoto University, Sakyo-ku, Kyoto 606-8502, Japan. E-mail: [email protected], [email protected] b. Department of Applied Biological Chemistry, Faculty of Agriculture, and Graduate School of Science and Technology, Niigata University, 8050 Ikarashi-2, Niigata 950-2181, Japan. E-mail: [email protected] c. Graduate School of Science, Osaka City University, 3-3-138 Sugimoto, Sumiyoshi, Osaka 558-8585, Japan. S1 Table of Contents Experimental Procedures S3 General procedure S3 Vector construction, expression and purification of BalTS S3 Crystallization and crystallographic analysis of balTS S3 Oligomeric state analysis S4 Analysis of the dimer geometry S4 Vector construction, expression and purification of BsuTS S4 Homology modeling of BsuTS S4 Enzymatic assays S5 Isolation and identification of β-springen S5 Movie Captions S6 Author Contributions S6 Figures Fig. S1 S7 Fig. S2 S8 Fig. S3 S9 Fig. S4 S10 Fig. S5 S17 Fig. S6 S18 Fig. S7, S8 S19 Fig. S9 S20 Fig. S10 S21 Fig. S11 S22 Tables Table S1 S23 Table S2 S24 References S27 S2 Experimental Procedures General procedure NMR spectra were recorded using a Bruker DPX 400 spectrometer at 400 MHz for proton (1H) and 100 MHz for carbon (13C). -

Distribution of Epiphytic Macroalgae on the Thalli of Their Hosts in Cuba
Acta Botanica Brasilica 27(4): 815-826. 2013. Distribution of epiphytic macroalgae on the thalli of their hosts in Cuba Yander Luis Diez García1, Abdiel Jover Capote2,6, Ana María Suárez Alfonso3, Liliana María Gómez Luna4 and Mutue Toyota Fujii5 Received: 4 April, 2012. Accepted: 14 October, 2013 ABSTRACT We investigated the distribution of epiphytic macroalgae on the thalli of their hosts at eight localities along the sou- theastern coast of Cuba between June 2010 and March 2011. We divided he epiphytes in two groups according to their distribution on the host: those at the base of the thallus and those on its surface. We determining the dissimilarity between the zones and the species involved. We identified 102 taxa of epiphytic macroalgae. There were significant differences between the two zones. In 31 hosts, the number of epiphytes was higher on the surface of the thallus, whereas the number of epiphytes was higher at the thallus base in 25 hosts, and the epiphytes were equally distributed between the two zones in five hosts (R=−0.001, p=0.398). The mean dissimilarity between the two zones, in terms of the species composition of the epiphytic macroalgae, was 96.64%. Hydrolithon farinosum and Polysiphonia atlantica accounted for 43.76% of the dissimilarity. Among macroalgae, the structure of the thallus seems to be a determinant of their viability as hosts for epiphytes. Key words: Chlorophyta, epiphytism, distribution, Phaeophyceae, Rhodophyta Introduction scales is a potentially productive approach. It is important to understand the patterns of abundance of all sympatric The structure of intertidal marine communities is deter- epiphytic species along the various gradients, because in- mined by a combination of physical factors and biotic interac- terspecific relationships could represent one of the factors tions (Little & Kitching 1996; Wernberg & Connell 2008). -

The Genus "Laurencia (Rhodomelaceae
- Courier Forsch.-Inst. Senckenberg, 159: 113- 117 - Frankfurt a.M., 0 1.07.1993 The Genus Laurencia (Rhodomelaceae, Rhodophyta) in the Canary Islands I Figure Tha riamiir 1 ~i..-ni.-:~ 1 i.inwinnrtv :e a mrnian nr iii~~GUUJ uuursi~,iu knmvunvun a= a fiiuup vk Ir! !he A!!ari.!ic Cous, STUTC(!W) ?i?ade a short medium-sized, erect, fleshy or cartilaginous, red al- review of three typical European species: L. obtusa gae distributed from temperate to tropical waters. (HUDSON)LAMOUROUX, L. pinnatifida (HUDSON)LA- During the last few years severa1 collections along MoURoUX and L. hybrida (Dc.) LENORMAND.Other the coasts of the Canary Islands have shown the im- researches carried out in this troublesome genus in portant role of the Laurencia species in the intertidal the Atlantic Ocean were made by TAYLOR(1960) in communities; however, it is rather problematic to Eastern Tropical and Subtropical Coasts of America, identify rnany of the taxa observed, and it seems OLIVEIRA-FILHO(1 969) jn Brazil, MACNE(1 980) in necessary to make a biosystematic review of this ge- the French Atlantic Coasts, LAWSON& JOHN(1982) nus in the Macaronesian Region. in the West Coast of Africa, RODRICUEZDE RIOS LAMOUROUXin 1813 established the genus with 8 (1981), RODRIGUEZDERroS & SNTO (1982, 1985) and species, but he didn't mention a type species. Critica1 RODRIGUEZDE RIOS& LOBO(1984) in Venezuela. systematic studies have been made by several authors, C. ACAIU)H(1823, 1824), J.AGARDH(1842, 1851, 1880), DE TON1 (1903, 1924). YMADA(1931). after reviewing many type specimens from different American and European Herbaria.