Multiplexed Cancer Immune Response Analysis Ncounter® Pancancer Immune Profiling Panel for Gene Expression
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Labinvest201436.Pdf
472A ANNUAL MEETING ABSTRACTS in hepatic fi brosis despite reversal of hyperbilirubinemia with Omegaven©. Detailed is expensive, time-consuming and diffi cult for routine application. For these reasons, pathologic fi ndings of whole explanted native livers in children who have undergone we evaluated the potential role of immunohistochemistry (IHC) as a screening tool to a combined liver-intestine transplant on Omegaven therapy have not been reported. identify candidate cases for FISH analysis and for ALK inhibitor therapy in NSCLC. Design: Explanted whole native livers from seven patients who have received To verify the mRNA expression of EML4-ALK, we used a MassARRAY method in Omegaven© emulsion as parenteral nutrition therapy in the pretransplant period were a small subset of patients. evaluated. Histological features including degree of infl ammation, fi brosis, steatosis, Design: We performed FISH and IHC for ALK and mutational analysis for EGFR and cholestasis and portal bile duct proliferation were semiquantitatively scored according K-ras in 523 NSCLC specimens. We conducted IHC analysis with the monoclonal to established criteria. antibody D5F3 and a highly sensitive detection system. We also performed a Results: The pathological features of the seven native liver explants are summarized MassARRAY-based analysis in a small subset of 11 samples to detect EML4-ALK in the following table. rearrangement. Features Pt. 1 Pt. 2 Pt. 3 Pt. 4 Pt. 5 Pt. 6 Pt. 7 Results: Of the 523 NSCLC specimens, 20 (3.8%) were positive for ALK rearrangement Protal infl ammation(0-4) 1111121 by FISH analysis. EGFR and Kras mutations were identifi ed in 70 (13.4%) and 124 Bile Duct Proliferation(0-3) 1311131 (23.7%) out of 523 tumor samples, respectively. -

Cancer Immunity (1 December 2006) Vol
Cancer Immun 1424-9634Academy of Cancer Immunology Cancer Immunity (1 December 2006) Vol. 6, p. 12 Submitted: 26 September 2006. Accepted: 10 October 2006. Copyright © 2006 by Andrew J. G. Simpson 061012 Article Physical interaction of two cancer-testis antigens, MAGE-C1 (CT7) and NY-ESO-1 (CT6) Hearn J. Cho1*,**, Otavia L. Caballero2*, Sacha Gnjatic2, Valéria C. C. Andrade3, Gisele W. Colleoni3, Andre L. Vettore4, Hasina H. Outtz1, Sheila Fortunato2, Nasser Altorki1, Cathy A. Ferrera1, Ramon Chua2, Achim A. Jungbluth2, Yao-Tseng Chen1, Lloyd J. Old2 and Andrew J. G. Simpson2 1Weill Medical College of Cornell University, 1300 York Avenue, New York, NY 10021, USA 2Ludwig Institute for Cancer Research, New York Branch at Memorial Sloan-Kettering Cancer Center, 1275 York Avenue, New York, NY 10021, USA 3Escola Paulista de Medicina, Universidade Federal de Sao Paulo, Sao Paulo, SP, Brazil 4Ludwig Institute for Cancer Research, Sao Paulo Branch, Sao Paulo, SP, Brazil *These authors contributed equally to this work **Present address: NYU Cancer Institute, New York University School of Medicine, 550 First Avenue, New York, NY 10016, USA Contributed by: LJ Old Cancer/testis (CT) antigens are the protein products of germ line- encoded on the X chromosome (CT-X antigens) and those that associated genes that are activated in a wide variety of tumors and can are not (non-X CT antigens) (1). elicit autologous cellular and humoral immune responses. CT It is estimated that 10% of the genes on the X-chromosome antigens can be divided between those that are encoded on the X belong to CT-X families (5). -

Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse
Welcome to More Choice CD Marker Handbook For more information, please visit: Human bdbiosciences.com/eu/go/humancdmarkers Mouse bdbiosciences.com/eu/go/mousecdmarkers Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse CD3 CD3 CD (cluster of differentiation) molecules are cell surface markers T Cell CD4 CD4 useful for the identification and characterization of leukocytes. The CD CD8 CD8 nomenclature was developed and is maintained through the HLDA (Human Leukocyte Differentiation Antigens) workshop started in 1982. CD45R/B220 CD19 CD19 The goal is to provide standardization of monoclonal antibodies to B Cell CD20 CD22 (B cell activation marker) human antigens across laboratories. To characterize or “workshop” the antibodies, multiple laboratories carry out blind analyses of antibodies. These results independently validate antibody specificity. CD11c CD11c Dendritic Cell CD123 CD123 While the CD nomenclature has been developed for use with human antigens, it is applied to corresponding mouse antigens as well as antigens from other species. However, the mouse and other species NK Cell CD56 CD335 (NKp46) antibodies are not tested by HLDA. Human CD markers were reviewed by the HLDA. New CD markers Stem Cell/ CD34 CD34 were established at the HLDA9 meeting held in Barcelona in 2010. For Precursor hematopoetic stem cell only hematopoetic stem cell only additional information and CD markers please visit www.hcdm.org. Macrophage/ CD14 CD11b/ Mac-1 Monocyte CD33 Ly-71 (F4/80) CD66b Granulocyte CD66b Gr-1/Ly6G Ly6C CD41 CD41 CD61 (Integrin b3) CD61 Platelet CD9 CD62 CD62P (activated platelets) CD235a CD235a Erythrocyte Ter-119 CD146 MECA-32 CD106 CD146 Endothelial Cell CD31 CD62E (activated endothelial cells) Epithelial Cell CD236 CD326 (EPCAM1) For Research Use Only. -

Complementary DNA Microarray Analysis of Chemokines and Their Receptors in Allergic Rhinitis RX Zhang,1 SQ Yu,2 JZ Jiang,3 GJ Liu3
RX Zhang, et al ORIGINAL ARTICLE Complementary DNA Microarray Analysis of Chemokines and Their Receptors in Allergic Rhinitis RX Zhang,1 SQ Yu,2 JZ Jiang,3 GJ Liu3 1 Department of Otolaryngology, Huadong Hospital, Fudan University, Shanghai, China 2 Department of Otolaryngology , Jinan General Hospital of PLA, Shandong, China 3 Department of Otolaryngology, Changhai Hospital, Second Military Medical University, Shanghai, China ■ Abstract Objective: To analyze the roles of chemokines and their receptors in the pathogenesis of allergic rhinitis by observing the complementary DNA (cDNA) expression of the chemokines and their receptors in the nasal mucosa of patients with and without allergic rhinitis, using gene chips. Methods: The total RNAs were isolated from the nasal mucosa of 20 allergic rhinitis patients and purifi ed to messenger RNAs, and then reversely transcribed to cDNAs and incorporated with samples of fl uorescence-labeled with Cy5-dUPT (rhinitis patient samples) or Cy3- dUTP (control samples of nonallergic nasal mucosa). Thirty-nine cDNAs of chemokines and their receptors were latticed into expression profi le chips, which were hybridized with probes and then scanned with the computer to study gene expression according to the different fl uorescence intensities. Results: The cDNAs of the following chemokines were upregulated: CCL1, CCL2, CCL5, CCL7, CCL8, CCL11, CCL13, CCL14, CCL17, CCL18, CCL19, CCL24, and CX3CL1 in most of the allergic rhinitis sample chips. CCR2, CCR3, CCR4, CCR5, CCR8 and CX3CR1 were the highly expressed receptor genes. Low expression of CXCL4 was found in these tissues. Conclusion: The T helper cell (TH) immune system is not well regulated in allergic rhinitis. -

Anti-CCL13 / MCP4 Antibody (Biotin) (ARG65993)
Product datasheet [email protected] ARG65993 Package: 50 μg anti-CCL13 / MCP4 antibody (Biotin) Store at: 4°C Summary Product Description Biotin-conjugated Goat Polyclonal antibody recognizes CCL13 / MCP4 Tested Reactivity Hu Tested Application ELISA, WB Host Goat Clonality Polyclonal Isotype IgG Target Name CCL13 / MCP4 Antigen Species Human Immunogen E. coli derived recombinant Human CCL13 / MCP4. (QPDALNVPST CCFTFSSKKI SLQRLKSYVI TTSRCPQKAV IFRTKLGKEI CADPKEKWVQ NYMKHLGRKA HTLKT) Conjugation Biotin Alternate Names SCYA13; C-C motif chemokine 13; Monocyte chemotactic protein 4; Small-inducible cytokine A13; CKb10; SCYL1; Monocyte chemoattractant protein 4; MCP-4; NCC-1; NCC1; CK-beta-10 Application Instructions Application table Application Dilution ELISA Direct: 0.25 - 1.0 µg/ml Sandwich: 0.25 - 1.0 µg/ml with ARG65992 as a capture antibody WB 0.1 - 0.2 µg/ml Application Note * The dilutions indicate recommended starting dilutions and the optimal dilutions or concentrations should be determined by the scientist. Calculated Mw 11 kDa Properties Form Liquid Purification Purified by affinity chromatography. Buffer PBS (pH 7.2) Concentration 1 mg/ml Storage instruction Aliquot and store in the dark at 2-8°C. Keep protected from prolonged exposure to light. Avoid repeated freeze/thaw cycles. Suggest spin the vial prior to opening. The antibody solution should be gently mixed before use. Note For laboratory research only, not for drug, diagnostic or other use. www.arigobio.com 1/2 Bioinformation Database links GeneID: 6357 Human Swiss-port # Q99616 Human Gene Symbol CCL13 Gene Full Name chemokine (C-C motif) ligand 13 Background This antimicrobial gene is one of several Cys-Cys (CC) cytokine genes clustered on the q-arm of chromosome 17. -
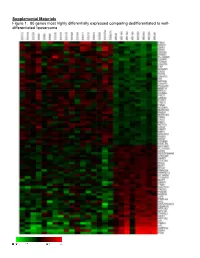
Supplemental Materials Figure 1. 80 Genes Most Highly Differentially Expressed Comparing Dedifferentiated to Well- Differentiated Liposarcoma
Supplemental Materials Figure 1. 80 genes most highly differentially expressed comparing dedifferentiated to well- differentiated liposarcoma Figure 2. Validation of microa CDC2, RACGAP1, FGFR-2, MAD2 and CITED1 200 CDK4 by Liposarcoma Subtype 16 0 12 0 80 Expression40 Level 0 Nl Fat rray data by quantitative RT-P Well Diff 16 0 p16 by Liposarcoma Subtype Dediff 12 0 80 Myxoid Expression40 Level Round Cell 0 12 0 MDM2 by Liposarcoma Subtype 10 0 Pleomorphic Nl Fat 80 60 40 Expression Level 20 RACGAP1 by LiposarcomaWell Diff Subtype 0 70 CR for genes CDK4, MDM2, p16, 60 Dediff 50 Nl Fat 40 30 Myxoid Expression20 Level Well Diff 10 50 0 Round Cell CDC2 by Liposarcoma Subtype Dediff 40 Nl Fat Pleomorphic 30 Myxoid 20 Expression Level Well Diff 10 MAD2L1 by Liposarcoma Subtype Round Cell 60 0 50 Dediff Pleomorphic 40 Nl Fat 30 Myxoid 20 Expression Level Well Diff 10 FGFR-2 by Liposarcoma Subtype 0 Round Cell 200 16 0 Dediff Nl Fat Pleomorphic 12 0 Myxoid 80 Expression Level Well Diff 40 Round Cell 0 Dediff Pleomorphic Nl Fat Myxoid Well Diff CITED1 by Liposarcoma Subtype Round Cell 10 0 80 Dediff Pleomorphic 60 40 Myxoid Expression Level 20 0 Round Cell Nl Fat Pleomorphic Well Diff Dediff Myxoid Round Cell Pleomorphic Table 1. 142 Gene classifier for liposarcoma subtype The genes used for each pair wise subtype comparison are grouped together. The flag column indicates which genes are unique to each subtype comparison. The values show the mean expression levels (actually the mean of the log expression levels was computed and than transformed back to absolute expression level). -

The Inactive X Chromosome Is Epigenetically Unstable and Transcriptionally Labile in Breast Cancer
Supplemental Information The inactive X chromosome is epigenetically unstable and transcriptionally labile in breast cancer Ronan Chaligné1,2,3,8, Tatiana Popova1,4, Marco-Antonio Mendoza-Parra5, Mohamed-Ashick M. Saleem5 , David Gentien1,6, Kristen Ban1,2,3,8, Tristan Piolot1,7, Olivier Leroy1,7, Odette Mariani6, Hinrich Gronemeyer*5, Anne Vincent-Salomon*1,4,6,8, Marc-Henri Stern*1,4,6 and Edith Heard*1,2,3,8 Extended Experimental Procedures Cell Culture Human Mammary Epithelial Cells (HMEC, Invitrogen) were grown in serum-free medium (HuMEC, Invitrogen). WI- 38, ZR-75-1, SK-BR-3 and MDA-MB-436 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen) containing 10% fetal bovine serum (FBS). DNA Methylation analysis. We bisulfite-treated 2 µg of genomic DNA using Epitect bisulfite kit (Qiagen). Bisulfite converted DNA was amplified with bisulfite primers listed in Table S3. All primers incorporated a T7 promoter tag, and PCR conditions are available upon request. We analyzed PCR products by MALDI-TOF mass spectrometry after in vitro transcription and specific cleavage (EpiTYPER by Sequenom®). For each amplicon, we analyzed two independent DNA samples and several CG sites in the CpG Island. Design of primers and selection of best promoter region to assess (approx. 500 bp) were done by a combination of UCSC Genome Browser (http://genome.ucsc.edu) and MethPrimer (http://www.urogene.org). All the primers used are listed (Table S3). NB: MAGEC2 CpG analysis have been done with a combination of two CpG island identified in the gene core. Analysis of RNA allelic expression profiles (based on Human SNP Array 6.0) DNA and RNA hybridizations were normalized by Genotyping console. -

Interleukin (IL)17A, F and AF in Inflammation: a Study in Collageninduced Arthritis and Rheumatoid Arthritis
bs_bs_banner Clinical and Experimental Immunology ORIGINAL ARTICLE doi:10.1111/cei.12376 Interleukin (IL)-17A, F and AF in inflammation: a study in collagen-induced arthritis and rheumatoid arthritis S. Sarkar,* S. Justa,* M. Brucks,* Summary J. Endres,† D. A. Fox,† X. Zhou,* Interleukin (IL)-17 plays a critical role in inflammation. Most studies to date F. Alnaimat,* B. Whitaker,‡ have elucidated the inflammatory role of IL-17A, often referred to as IL-17. J. C. Wheeler,‡ B. H. Jones§ and IL-17F is a member of the IL-17 family bearing 50% homology to IL-17A S. R. Bommireddy* and can also be present as heterodimer IL-17AF. This study elucidates the *Section of Rheumatology, Department of Medicine, and the Arizona Arthritis Center, distribution and contribution of IL-17A, F and AF in inflammatory arthritis. University of Arizona, Tucson, AZ, †Divison of Neutralizing antibody to IL-17A alone or IL-17F alone or in combination Rheumatology, Department of Internal Medicine, was utilized in the mouse collagen-induced arthritis (CIA) model to eluci- University of Michigan, Ann Arbor, MI, and date the contribution of each subtype in mediating inflammation. IL-17A, F ‡Biologics Research and §Immunology Discovery and AF were all increased during inflammatory arthritis. Neutralization of Research, Janssen Research and Development, IL-17A reduced the severity of arthritis, neutralization of IL-17A+IL-17F had Spring House, PA, USA the same effect as neutralizing IL-17A, while neutralization of IL-17F had no effect. Moreover, significantly higher levels of IL-17A and IL-17F were detected in peripheral blood mononuclear cells (PBMC) from patients with rheumatoid arthritis (RA) in comparison to patients with osteoarthritis (OA). -

Inflammation-Dependent IL18 Signaling Restricts Hepatocellular Carcinoma Growth by Enhancing the Accumulation and Activity of Tumor-Infiltrating Lymphocytes
Published OnlineFirst February 18, 2016; DOI: 10.1158/0008-5472.CAN-15-1548 Cancer Tumor and Stem Cell Biology Research Inflammation-Dependent IL18 Signaling Restricts Hepatocellular Carcinoma Growth by Enhancing the Accumulation and Activity of Tumor- Infiltrating Lymphocytes Geoffrey J. Markowitz1, Pengyuan Yang1,2,3, Jing Fu3, Gregory A. Michelotti4, Rui Chen1, Jianhua Sui5, Bin Yang2, Wen-Hao Qin3, Zheng Zhang6, Fu-Sheng Wang6, Anna Mae Diehl4, Qi-Jing Li7, Hongyang Wang3, and Xiao-Fan Wang1 Abstract Chronic inflammation in liver tissue is an underlying cause of IL18R1 deletion increased tumor burden. Mechanistically, we hepatocellular carcinoma. High levels of inflammatory cytokine foundthatIL18exertedinflammation-dependent tumor-sup- IL18 in the circulation of patients with hepatocellular carcinoma pressive effects largely by promoting the differentiation, activ- correlates with poor prognosis. However, conflicting results have ity, and survival of tumor-infiltrating T cells. Finally, differences been reported for IL18 in hepatocellular carcinoma development in the expression of IL18 in tumor tissue versus nontumor and progression. In this study, we used tissue specimens from tissueweremorepredictiveofpatientoutcomethanoverall hepatocellular carcinoma patients and clinically relevant mouse tissue expression. Taken together, our findings resolve a long- models of hepatocellular carcinoma to evaluate IL18 expression standing contradiction regarding a tumor-suppressive role for and function. In a mouse model of liver fibrosis that recapitulates IL18 in established hepatocellular carcinoma and provide a a tumor-promoting microenvironment, global deletion of the mechanistic explanation for the complex relationship between IL18 receptor IL18R1 enhanced tumor growth and burden. Sim- its expression pattern and hepatocellular carcinoma prognosis. ilarly, in a carcinogen-induced model of liver tumorigenesis, Cancer Res; 76(8); 1–12. -

Bioinformatics Identification of CCL8/21 As Potential Prognostic
Bioscience Reports (2020) 40 BSR20202042 https://doi.org/10.1042/BSR20202042 Research Article Bioinformatics identification of CCL8/21 as potential prognostic biomarkers in breast cancer microenvironment 1,* 2,* 3 4 5 1 Bowen Chen , Shuyuan Zhang ,QiuyuLi, Shiting Wu ,HanHe and Jinbo Huang Downloaded from http://portlandpress.com/bioscirep/article-pdf/40/11/BSR20202042/897847/bsr-2020-2042.pdf by guest on 28 September 2021 1Department of Breast Disease, Maoming People’s Hospital, Maoming 525000, China; 2Department of Clinical Laboratory, Maoming People’s Hospital, Maoming 525000, China; 3Department of Emergency, Maoming People’s Hospital, Maoming 525000, China; 4Department of Oncology, Maoming People’s Hospital, Maoming 525000, China; 5Department of Medical Imaging, Maoming People’s Hospital, Maoming 525000, China Correspondence: Shuyuan Zhang ([email protected]) Background: Breast cancer (BC) is the most common malignancy among females world- wide. The tumor microenvironment usually prevents effective lymphocyte activation and infiltration, and suppresses infiltrating effector cells, leading to a failure of the host toreject the tumor. CC chemokines play a significant role in inflammation and infection. Methods: In our study, we analyzed the expression and survival data of CC chemokines in patients with BC using several bioinformatics analyses tools. Results: The mRNA expression of CCL2/3/4/5/7/8/11/17/19/20/22 was remark- ably increased while CCL14/21/23/28 was significantly down-regulated in BC tis- sues compared with normal tissues. Methylation could down-regulate expression of CCL2/5/15/17/19/20/22/23/24/25/26/27 in BC. Low expression of CCL3/4/23 was found to be associated with drug resistance in BC. -

Supplementary Table 1: Adhesion Genes Data Set
Supplementary Table 1: Adhesion genes data set PROBE Entrez Gene ID Celera Gene ID Gene_Symbol Gene_Name 160832 1 hCG201364.3 A1BG alpha-1-B glycoprotein 223658 1 hCG201364.3 A1BG alpha-1-B glycoprotein 212988 102 hCG40040.3 ADAM10 ADAM metallopeptidase domain 10 133411 4185 hCG28232.2 ADAM11 ADAM metallopeptidase domain 11 110695 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 195222 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 165344 8751 hCG20021.3 ADAM15 ADAM metallopeptidase domain 15 (metargidin) 189065 6868 null ADAM17 ADAM metallopeptidase domain 17 (tumor necrosis factor, alpha, converting enzyme) 108119 8728 hCG15398.4 ADAM19 ADAM metallopeptidase domain 19 (meltrin beta) 117763 8748 hCG20675.3 ADAM20 ADAM metallopeptidase domain 20 126448 8747 hCG1785634.2 ADAM21 ADAM metallopeptidase domain 21 208981 8747 hCG1785634.2|hCG2042897 ADAM21 ADAM metallopeptidase domain 21 180903 53616 hCG17212.4 ADAM22 ADAM metallopeptidase domain 22 177272 8745 hCG1811623.1 ADAM23 ADAM metallopeptidase domain 23 102384 10863 hCG1818505.1 ADAM28 ADAM metallopeptidase domain 28 119968 11086 hCG1786734.2 ADAM29 ADAM metallopeptidase domain 29 205542 11085 hCG1997196.1 ADAM30 ADAM metallopeptidase domain 30 148417 80332 hCG39255.4 ADAM33 ADAM metallopeptidase domain 33 140492 8756 hCG1789002.2 ADAM7 ADAM metallopeptidase domain 7 122603 101 hCG1816947.1 ADAM8 ADAM metallopeptidase domain 8 183965 8754 hCG1996391 ADAM9 ADAM metallopeptidase domain 9 (meltrin gamma) 129974 27299 hCG15447.3 ADAMDEC1 ADAM-like, -

Cytokine-Enhanced Cytolytic Activity of Exosomes from NK Cells
Cancer Gene Therapy https://doi.org/10.1038/s41417-021-00352-2 ARTICLE Cytokine-enhanced cytolytic activity of exosomes from NK Cells 1 1 2 3 2 3 Yutaka Enomoto ● Peng Li ● Lisa M. Jenkins ● Dimitrios Anastasakis ● Gaelyn C. Lyons ● Markus Hafner ● Warren J. Leonard 1 Received: 4 February 2021 / Revised: 9 May 2021 / Accepted: 18 May 2021 This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply 2021. This article is published with open access Abstract Natural killer (NK) cells play key roles in immune surveillance against tumors and viral infection. NK cells distinguish abnormal cells from healthy cells by cell–cell interaction with cell surface proteins and then attack target cells via multiple mechanisms. In addition, extracellular vesicles (EVs) derived from NK cells (NK-EVs), including exosomes, possess cytotoxic capacity against tumor cells, but their characteristics and regulation by cytokines remain unknown. Here, we report that EVs derived from human NK-92 cells stimulated with IL-15 + IL-21 show enhanced cytotoxic capacity against tumor cells. Major cytolytic granules, granzyme B and granzyme H, are enriched by IL-15 + IL-21 stimulation in NK-EVs; however, knockout experiments reveal those cytolytic granules are independent of enhanced cytotoxic capacity. To find out the key molecules, mass spectrometry analyses were 1234567890();,: 1234567890();,: performed with different cytokine conditions, no cytokine, IL-15, IL-21, or IL-15 + IL-21. We then found that CD226 (DNAM-1) on NK-EVs is enriched by IL-15 + IL-21 stimulation and that blocking antibodies against CD226 reduced the cytolytic activity of NK-EVs.