WO 2014/068408 A2 8 May 2014 (08.05.2014) P O P C T
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Exome Sequencing Identifies Mutations in the Gene TTC7A In
Developmental defects J Med Genet: first published as 10.1136/jmedgenet-2012-101483 on 19 February 2013. Downloaded from ORIGINAL ARTICLE Exome sequencing identifies mutations in the gene TTC7A in French-Canadian cases with hereditary multiple intestinal atresia Mark E Samuels,1 Jacek Majewski,2 Najmeh Alirezaie,2 Isabel Fernandez,1,3 Ferran Casals,1 Natalie Patey,1,4 Hélène Decaluwe,1,5 Isabelle Gosselin,6 Elie Haddad,1,3,5 Alan Hodgkinson,1 Youssef Idaghdour,1 Valerie Marchand,1,5 1,5 6,7 6 6 Open Access Jacques L Michaud, Marc-André Rodrigue, Sylvie Desjardins, Stéphane Dubois, Scan to access more 1,3 1,5 6,7 8 free content Francoise Le Deist, Philip Awadalla, Vincent Raymond, Bruno Maranda ▸ Additional material is ABSTRACT attempted, outcomes are poor and the condition is published online only. To view Background Congenital multiple intestinal atresia usual fatal within the first month of life. To date, please visit the journal online (http://dx.doi.org/10.1136/ (MIA) is a severe, fatal neonatal disorder, involving the no primary aetiology has been proved for the con- jmedgenet-2012-101483). occurrence of obstructions in the small and large dition. Importantly, in some cases MIA is also asso- 1 intestines ultimately leading to organ failure. Surgical ciated with either mild or severe combined Centre de Recherche du CHU fi 2–5 Ste-Justine, University of interventions are palliative but do not provide long-term immunode ciency (SCID), raising the possibility Montreal, Montreal, Quebec, survival. Severe immunodeficiency may be associated that an abnormal immune response might be the Canada with the phenotype. -

Differences Between Human and Chimpanzee Genomes and Their Implications in Gene Expression, Protein Functions and Biochemical Properties of the Two Species Maria V
Suntsova and Buzdin BMC Genomics 2020, 21(Suppl 7):535 https://doi.org/10.1186/s12864-020-06962-8 REVIEW Open Access Differences between human and chimpanzee genomes and their implications in gene expression, protein functions and biochemical properties of the two species Maria V. Suntsova1 and Anton A. Buzdin1,2,3,4* From 11th International Young Scientists School “Systems Biology and Bioinformatics”–SBB-2019 Novosibirsk, Russia. 24-28 June 2019 Abstract Chimpanzees are the closest living relatives of humans. The divergence between human and chimpanzee ancestors dates to approximately 6,5–7,5 million years ago. Genetic features distinguishing us from chimpanzees and making us humans are still of a great interest. After divergence of their ancestor lineages, human and chimpanzee genomes underwent multiple changes including single nucleotide substitutions, deletions and duplications of DNA fragments of different size, insertion of transposable elements and chromosomal rearrangements. Human-specific single nucleotide alterations constituted 1.23% of human DNA, whereas more extended deletions and insertions cover ~ 3% of our genome. Moreover, much higher proportion is made by differential chromosomal inversions and translocations comprising several megabase-long regions or even whole chromosomes. However, despite of extensive knowledge of structural genomic changes accompanying human evolution we still cannot identify with certainty the causative genes of human identity. Most structural gene-influential changes happened at the level of expression regulation, which in turn provoked larger alterations of interactome gene regulation networks. In this review, we summarized the available information about genetic differences between humans and chimpanzees and their potential functional impacts on differential molecular, anatomical, physiological and cognitive peculiarities of these species. -
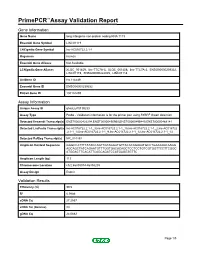
Primepcr™Assay Validation Report
PrimePCR™Assay Validation Report Gene Information Gene Name long intergenic non-protein coding RNA 1119 Ensembl Gene Symbol LINC01119 LNCipedia Gene Symbol lnc-AC016722.2.1-1 Organism Human Ensembl Gene Aliases Not Available LCNipedia Gene Aliases XLOC_001459, linc-TTC7A-3, XLOC_001458, linc-TTC7A-2, ENSG00000239332, LINC01119, ENSG00000222005, LINC01118 UniGene ID Hs.114449 Ensembl Gene ID ENSG00000239332 Entrez Gene ID 100134259 Assay Information Unique Assay ID qhsaLEP0139233 Assay Type Probe - Validation information is for the primer pair using SYBR® Green detection Detected Ensembl Transcript(s) ENST00000422294,ENST00000490950,ENST00000495449,ENST00000468141 Detected LncPedia Transcript(s) lnc-AC016722.2.1-1_3,lnc-AC016722.2.1-1_15,lnc-AC016722.2.1-1_2,lnc-AC016722 .2.1-1_14,lnc-AC016722.2.1-1_9,lnc-AC016722.2.1-1_12,lnc-AC016722.2.1-1_13 Detected RefSeq Transcript(s) NR_024452 Amplicon Context Sequence GGGCCATTTTATACCAGTTGTAGGATGTTACACAGAAATGCCTGAAAAGCAAGG AGCAGCTATCAGAATGTTTGGTGACACAGCTCCTCCTGTCGTGGTTCCTTCGGC ATGGACTTCACATTCAGCAGATCCATGAGGTGTTC Amplicon Length (bp) 113 Chromosome Location chr2:46855974-46858205 Assay Design Exonic Validation Results Efficiency (%) 99.5 R2 0.9986 cDNA Cq 27.2537 cDNA Tm (Celsius) 83 gDNA Cq 24.0682 Page 1/5 PrimePCR™Assay Validation Report Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 2/5 PrimePCR™Assay Validation Report LINC01119, Human Amplification Plot Amplification of cDNA generated from 25 ng of universal reference RNA Melt Peak -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

The Capacity of Long-Term in Vitro Proliferation of Acute Myeloid
The Capacity of Long-Term in Vitro Proliferation of Acute Myeloid Leukemia Cells Supported Only by Exogenous Cytokines Is Associated with a Patient Subset with Adverse Outcome Annette K. Brenner, Elise Aasebø, Maria Hernandez-Valladares, Frode Selheim, Frode Berven, Ida-Sofie Grønningsæter, Sushma Bartaula-Brevik and Øystein Bruserud Supplementary Material S2 of S31 Table S1. Detailed information about the 68 AML patients included in the study. # of blasts Viability Proliferation Cytokine Viable cells Change in ID Gender Age Etiology FAB Cytogenetics Mutations CD34 Colonies (109/L) (%) 48 h (cpm) secretion (106) 5 weeks phenotype 1 M 42 de novo 241 M2 normal Flt3 pos 31.0 3848 low 0.24 7 yes 2 M 82 MF 12.4 M2 t(9;22) wt pos 81.6 74,686 low 1.43 969 yes 3 F 49 CML/relapse 149 M2 complex n.d. pos 26.2 3472 low 0.08 n.d. no 4 M 33 de novo 62.0 M2 normal wt pos 67.5 6206 low 0.08 6.5 no 5 M 71 relapse 91.0 M4 normal NPM1 pos 63.5 21,331 low 0.17 n.d. yes 6 M 83 de novo 109 M1 n.d. wt pos 19.1 8764 low 1.65 693 no 7 F 77 MDS 26.4 M1 normal wt pos 89.4 53,799 high 3.43 2746 no 8 M 46 de novo 26.9 M1 normal NPM1 n.d. n.d. 3472 low 1.56 n.d. no 9 M 68 MF 50.8 M4 normal D835 pos 69.4 1640 low 0.08 n.d. -

Learning from Cadherin Structures and Sequences: Affinity Determinants and Protein Architecture
Learning from cadherin structures and sequences: affinity determinants and protein architecture Klára Fels ıvályi Submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Graduate School of Arts and Sciences COLUMBIA UNIVERSITY 2014 © 2014 Klara Felsovalyi All rights reserved ABSTRACT Learning from cadherin structures and sequences: affinity determinants and protein architecture Klara Felsovalyi Cadherins are a family of cell-surface proteins mediating adhesion that are important in development and maintenance of tissues. The family is defined by the repeating cadherin domain (EC) in their extracellular region, but they are diverse in terms of protein size, architecture and cellular function. The best-understood subfamily is the type I classical cadherins, which are found in vertebrates and have five EC domains. Among the five different type I classical cadherins, the binding interactions are highly specific in their homo- and heterophilic binding affinities, though their sequences are very similar. As previously shown, E- and N-cadherins, two prototypic members of the subfamily, differ in their homophilic K D by about an order of magnitude, while their heterophilic affinity is intermediate. To examine the source of the binding affinity differences among type I cadherins, we used crystal structures, analytical ultracentrifugation (AUC), surface plasmon resonance (SPR), and electron paramagnetic resonance (EPR) studies. Phylogenetic analysis and binding affinity behavior show that the type I cadherins can be further divided into two subgroups, with E- and N-cadherin representing each. In addition to the affinity differences in their wild-type binding through the strand-swapped interface, a second interface also shows an affinity difference between E- and N-cadherin. -

Structural Basis of Sterol Recognition and Nonvesicular Transport by Lipid
Structural basis of sterol recognition and nonvesicular PNAS PLUS transport by lipid transfer proteins anchored at membrane contact sites Junsen Tonga, Mohammad Kawsar Manika, and Young Jun Ima,1 aCollege of Pharmacy, Chonnam National University, Bukgu, Gwangju, 61186, Republic of Korea Edited by David W. Russell, University of Texas Southwestern Medical Center, Dallas, TX, and approved December 18, 2017 (received for review November 11, 2017) Membrane contact sites (MCSs) in eukaryotic cells are hotspots for roidogenic acute regulatory protein-related lipid transfer), PITP lipid exchange, which is essential for many biological functions, (phosphatidylinositol/phosphatidylcholine transfer protein), Bet_v1 including regulation of membrane properties and protein trafficking. (major pollen allergen from birch Betula verrucosa), PRELI (pro- Lipid transfer proteins anchored at membrane contact sites (LAMs) teins of relevant evolutionary and lymphoid interest), and LAMs contain sterol-specific lipid transfer domains [StARkin domain (SD)] (LTPs anchored at membrane contact sites) (9). and multiple targeting modules to specific membrane organelles. Membrane contact sites (MCSs) are closely apposed regions in Elucidating the structural mechanisms of targeting and ligand which two organellar membranes are in close proximity, typically recognition by LAMs is important for understanding the interorga- within a distance of 30 nm (7). The ER, a major site of lipid bio- nelle communication and exchange at MCSs. Here, we determined synthesis, makes contact with almost all types of subcellular or- the crystal structures of the yeast Lam6 pleckstrin homology (PH)-like ganelles (10). Oxysterol-binding proteins, which are conserved domain and the SDs of Lam2 and Lam4 in the apo form and in from yeast to humans, are suggested to have a role in the di- complex with ergosterol. -

Distinct Antiviral Signatures Revealed by the Magnitude and Round of Influenza Virus Replication in Vivo
Distinct antiviral signatures revealed by the magnitude and round of influenza virus replication in vivo Louisa E. Sjaastada,b,1, Elizabeth J. Fayb,c,1, Jessica K. Fiegea,b, Marissa G. Macchiettod, Ian A. Stonea,b, Matthew W. Markmana,b, Steven Shend, and Ryan A. Langloisa,b,c,2 aDepartment of Microbiology and Immunology, University of Minnesota, Minneapolis, MN 55455; bCenter for Immunology, University of Minnesota, Minneapolis, MN 55455; cBiochemistry, Molecular Biology and Biophysics Graduate Program, University of Minnesota, Minneapolis, MN 55455; and dInstitute for Health Informatics, University of Minnesota, Minneapolis, MN 55455 Edited by Michael B. A. Oldstone, The Scripps Research Institute, La Jolla, CA, and approved August 8, 2018 (received for review May 9, 2018) Influenza virus has a broad cellular tropism in the respiratory tract. virus cannot spread; therefore, any differences in viral abun- Infected epithelial cells sense the infection and initiate an antiviral dance will be a direct result of replication intensity. Infection of response. To define the antiviral response at the earliest stages of mice revealed uninfected cells and cells with both low and high infection we used a series of single-cycle reporter viruses. These levels of virus replication. These populations exhibited unique viral probes demonstrated cells in vivo harbor a range in magni- ISG signatures, and this finding was corroborated through the tude of virus replication. Transcriptional profiling of cells support- use of a reporter virus capable of specifically detecting active ing different levels of replication revealed tiers of IFN-stimulated replication. This suggests that the antiviral response is tuned to gene expression. -

Plasma Cells in Vitro Generation of Long-Lived Human
Downloaded from http://www.jimmunol.org/ by guest on September 24, 2021 is online at: average * The Journal of Immunology , 32 of which you can access for free at: 2012; 189:5773-5785; Prepublished online 16 from submission to initial decision 4 weeks from acceptance to publication November 2012; doi: 10.4049/jimmunol.1103720 http://www.jimmunol.org/content/189/12/5773 In Vitro Generation of Long-lived Human Plasma Cells Mario Cocco, Sophie Stephenson, Matthew A. Care, Darren Newton, Nicholas A. Barnes, Adam Davison, Andy Rawstron, David R. Westhead, Gina M. Doody and Reuben M. Tooze J Immunol cites 65 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription http://www.jimmunol.org/content/suppl/2012/11/16/jimmunol.110372 0.DC1 This article http://www.jimmunol.org/content/189/12/5773.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2012 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 24, 2021. The Journal of Immunology In Vitro Generation of Long-lived Human Plasma Cells Mario Cocco,*,1 Sophie Stephenson,*,1 Matthew A. -
![H00010618-M02 規格 : [ 100 Ug ] List All](https://docslib.b-cdn.net/cover/5961/h00010618-m02-100-ug-list-all-1125961.webp)
H00010618-M02 規格 : [ 100 Ug ] List All
TGOLN2 monoclonal antibody (M02), clone 2F11 Catalog # : H00010618-M02 規格 : [ 100 ug ] List All Specification Application Image Product Mouse monoclonal antibody raised against a partial recombinant Western Blot (Recombinant protein) Description: TGOLN2. Immunohistochemistry Immunogen: TGOLN2 (NP_006455, 229 a.a. ~ 327 a.a) partial recombinant protein (Formalin/PFA-fixed paraffin- with GST tag. MW of the GST tag alone is 26 KDa. embedded sections) Sequence: QGPIDGPSKSGAEEQTSKDSPNKVVPEQPSRKDHSKPISNPSDNKELPK ADTNQLADKGKLSPHAFKTESGEETDLISPPQEEVKSSEPTEDVEPKEAE Host: Mouse Reactivity: Human enlarge Isotype: IgG2a Kappa Immunofluorescence Quality Control Antibody Reactive Against Recombinant Protein. Testing: enlarge Sandwich ELISA (Recombinant protein) Western Blot detection against Immunogen (36.63 KDa) . enlarge Storage Buffer: In 1x PBS, pH 7.4 ELISA Storage Store at -20°C or lower. Aliquot to avoid repeated freezing and thawing. Instruction: MSDS: Download Datasheet: Download Publication Reference 1. Phenylalanine Residues at the Carboxyl-terminus of the Herpes Simplex Virus Type-1 UL20 Membrane Protein Regulate Cytoplasmic Virion Envelopment and Infectious Virus Production. Charles AS, Chouljenko VN, Jambunathan N, Subramanian R, Mottram P, Kousoulas KGJ Virol. 2014 Apr 23. 2. Distinctive features of degenerating Purkinje cells in spinocerebellar ataxia type 31. Yoshida K, Asakawa M, Suzuki-Kouyama E, Tabata K, Shintaku M, Ikeda S, Oyanagi KNeuropathology. 2014 Jun;34(3):261-7. doi: 10.1111/neup.12090. Epub 2013 Dec 17. 3. Abnormal localization of leucine-rich repeat kinase 2 to the endosomal-lysosomal compartment in lewy body disease. Page 1 of 3 2016/5/22 Higashi S, Moore DJ, Yamamoto R, Minegishi M, Sato K, Togo T, Katsuse O, Uchikado H, Furukawa Y, Hino H, Kosaka K, Emson PC, Wada K, Dawson VL, Dawson TM, Arai H, Iseki E.J Neuropathol Exp Neurol. -

Ec5c1b8e28a54f8ed17a1c301b
www.clinsci.org Clinical Science (2010) 119, 265–272 (Printed in Great Britain) doi:10.1042/CS20100266 265 ACCELERATED PUBLICATION Overexpression of STARD3 in human monocyte/macrophages induces an anti-atherogenic lipid phenotype Faye BORTHWICK, Anne-Marie ALLEN, Janice M. TAYLOR and Annette GRAHAM Vascular Biology Group, Department of Biological and Biomedical Sciences, Glasgow Caledonian University, Glasgow G4 0BA, U.K. ABSTRACT Dysregulated macrophage cholesterol homoeostasis lies at the heart of early and developing atheroma, and removal of excess cholesterol from macrophage foam cells, by efficient transport mechanisms, is central to stabilization and regression of atherosclerotic lesions. The present study demonstrates that transient overexpression of STARD3 {START [StAR (steroidogenic acute regulatory protein)-related lipid transfer] domain 3; also known as MLN64 (metastatic lymph node 64)}, an endosomal cholesterol transporter and member of the ‘START’ family of lipid trafficking proteins, induces significant increases in macrophage ABCA1 (ATP-binding cassette transporter A1) mRNA and protein, enhances [3H]cholesterol efflux to apo (apolipoprotein) AI, and reduces biosynthesis of cholesterol, cholesteryl ester, fatty acids, triacylglycerol and phospholipids from [14C]acetate, compared with controls. Notably, overexpression of STARD3 prevents increases in cholesterol esterification in response to acetylated LDL (low-density lipoprotein), blocking cholesteryl ester deposition. Thus enhanced endosomal trafficking via STARD3 induces an anti- atherogenic macrophage lipid phenotype, positing a potentially therapeutic strategy. Clinical Science INTRODUCTION and stabilization, and can be orchestrated, at least in vitro, by ABC (ATP-binding cassette) lipid trans- Dysregulated macrophage cholesterol homoeostasis lies porters such as ABCA1, ABCG1 and ABCG4, and at the heart of early and developing atheroma, the apo (apolipoprotein) acceptors, such as apoAI and apoE principal cause of coronary heart disease. -

Table of Contents Table of Contents
Table of contents Table of contents................................................................................................................. 1 Summary ............................................................................................................................. 3 Abbreviations ...................................................................................................................... 4 Gene Symbols I - Incidentals .............................................................................................. 5 Gene symbols II - Target Gene Names ............................................................................... 6 1. Introduction........................................................................................................... 12 1.1. The Pathos of the Crab ..................................................................................... 12 1.2. Cancer in the Molecular Age - a genetic and epigenetic disease..................... 13 1.3. Tumoral evolution............................................................................................. 14 1.4. DNA repair systems .......................................................................................... 17 1.4.1. DNA Mismatch Repair.............................................................................. 18 1.4.2. Double-strand Break Repair..................................................................... 19 1.4.3. Direct Repair...........................................................................................