(TGFBR3)-Associated Membranous Nephropathy
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Transforming Growth Factor-Beta Family Members Are Regulated During Induced Luteolysis in Cattle
a https://doi.org/10.21451/1984-3143-AR2018-0146 Original Article Animal Reproduction Anim Reprod. 2019;16(4):829-837 Transforming growth factor-beta family members are regulated during induced luteolysis in cattle Cristina Sangoi Haas1, Monique Tomazele Rovani1 , Gustavo Freitas Ilha2 , Kalyne Bertolin2 , Juliana Germano Ferst2 , Alessandra Bridi2 , Vilceu Bordignon3 , Raj Duggavathi3, Alfredo Quites Antoniazzi2 , Paulo Bayard Dias Gonçalves2 , Bernardo Garziera Gasperin1* 1Universidade Federal de Pelotas, Departamento de Patologia Animal, Capão do Leão, RS, Brasil 2Universidade Federal de Santa Maria, Laboratório de Biotecnologia e Reprodução Animal, Santa Maria, RS, Brasil 3McGill University, Department of Animal Science, Sainte-Anne-de-Bellevue, QC, Canada Abstract cell death and tissue remodeling (Miyamoto et al., 2009; Skarzynski and Okuda, 2010; Shirasuna et al., 2012). The transforming growth factors beta (TGFβ) Despite its essential role in reproduction, the cellular and are local factors produced by ovarian cells which, after molecular mechanisms mediating CL regression are not binding to their receptors, regulate follicular deviation fully understood. It is well established that CLs are not fully and ovulation. However, their regulation and function responsive to PGF until day 5-6 after ovulation, whereas during corpus luteum (CL) regression has been poorly after days 15-17, in the absence of gestation, spontaneous investigated. The present study evaluated the mRNA luteolysis occurs. Thus, performing PGF treatment on day regulation of some TGFβ family ligands and their receptors 10 after ovulation represents an adequate model to study in the bovine CL during induced luteolysis in vivo. On day CL regression, because all the CLs are fully responsive to 10 of the estrous cycle, cows received an injection of PGF and still functional. -

TGF Beta Signaling Pathway 1 TGF Beta Signaling Pathway
TGF beta signaling pathway 1 TGF beta signaling pathway The Transforming growth factor beta (TGFβ) signaling pathway is involved in many cellular processes in both the adult organism and the developing embryo including cell growth, cell differentiation, apoptosis, cellular homeostasis and other cellular functions. In spite of the wide range of cellular processes that the TGFβ signaling pathway regulates, the process is relatively simple. TGFβ superfamily ligands bind to a type II receptor, which recruits and phosphorylates a type I receptor. The type I receptor then phosphorylates receptor-regulated SMADs (R-SMADs) which can now bind the coSMAD SMAD4. R-SMAD/coSMAD complexes accumulate in the nucleus where they act as transcription factors and participate in the regulation of target gene expression. Mechanism Ligand Binding The TGF Beta superfamily of ligands include: Bone morphogenetic proteins (BMPs), Growth and differentiation factors (GDFs), Anti-müllerian hormone (AMH), Activin, Nodal and TGFβ's[1] . Signalling begins with the binding of a TGF beta superfamily ligand to a TGF beta type II receptor. The type II receptor is a serine/threonine receptor kinase, which catalyses the phosphorylation of the Type I receptor. Each class of ligand binds to a specific type II receptor[2] .In mammals there are seven known type I receptors and five type II receptors[3] . There are three activins: Activin A, Activin B and Activin AB. Activins are involved in embryogenesis and osteogenesis. They also regulate many hormones including pituitary, gonadal and hypothalamic hormones as well as insulin. They are also nerve cell survival factors. The BMPs bind to the Bone morphogenetic protein receptor type-2 (BMPR2). -

6 Signaling and BMP Antagonist Noggin in Prostate Cancer
[CANCER RESEARCH 64, 8276–8284, November 15, 2004] Bone Morphogenetic Protein (BMP)-6 Signaling and BMP Antagonist Noggin in Prostate Cancer Dominik R. Haudenschild, Sabrina M. Palmer, Timothy A. Moseley, Zongbing You, and A. Hari Reddi Center for Tissue Regeneration and Repair, Department of Orthopedic Surgery, School of Medicine, University of California, Davis, Sacramento, California ABSTRACT antagonists has recently been discovered. These are secreted proteins that bind to BMPs and reduce their bioavailability for interactions It has been proposed that the osteoblastic nature of prostate cancer with the BMP receptors. Extracellular BMP antagonists include nog- skeletal metastases is due in part to elevated activity of bone morphoge- gin, follistatin, sclerostatin, chordin, DCR, BMPMER, cerberus, netic proteins (BMPs). BMPs are osteoinductive morphogens, and ele- vated expression of BMP-6 correlates with skeletal metastases of prostate gremlin, DAN, and others (refs. 11–16; reviewed in ref. 17). There are cancer. In this study, we investigated the expression levels of BMPs and several type I and type II receptors that bind to BMPs with different their modulators in prostate, using microarray analysis of cell cultures affinities. BMP activity is also regulated at the cell membrane level by and gene expression. Addition of exogenous BMP-6 to DU-145 prostate receptor antagonists such as BAMBI (18), which acts as a kinase- cancer cell cultures inhibited their growth by up-regulation of several deficient receptor. Intracellularly, the regulation of BMP activity at cyclin-dependent kinase inhibitors such as p21/CIP, p18, and p19. Expres- the signal transduction level is even more complex. There are inhib- sion of noggin, a BMP antagonist, was significantly up-regulated by itory Smads (Smad-6 and Smad-7), as well as inhibitors of inhibitory BMP-6 by microarray analysis and was confirmed by quantitative reverse Smads (AMSH and Arkadia). -

Fibroblasts from the Human Skin Dermo-Hypodermal Junction Are
cells Article Fibroblasts from the Human Skin Dermo-Hypodermal Junction are Distinct from Dermal Papillary and Reticular Fibroblasts and from Mesenchymal Stem Cells and Exhibit a Specific Molecular Profile Related to Extracellular Matrix Organization and Modeling Valérie Haydont 1,*, Véronique Neiveyans 1, Philippe Perez 1, Élodie Busson 2, 2 1, 3,4,5,6, , Jean-Jacques Lataillade , Daniel Asselineau y and Nicolas O. Fortunel y * 1 Advanced Research, L’Oréal Research and Innovation, 93600 Aulnay-sous-Bois, France; [email protected] (V.N.); [email protected] (P.P.); [email protected] (D.A.) 2 Department of Medical and Surgical Assistance to the Armed Forces, French Forces Biomedical Research Institute (IRBA), 91223 CEDEX Brétigny sur Orge, France; [email protected] (É.B.); [email protected] (J.-J.L.) 3 Laboratoire de Génomique et Radiobiologie de la Kératinopoïèse, Institut de Biologie François Jacob, CEA/DRF/IRCM, 91000 Evry, France 4 INSERM U967, 92260 Fontenay-aux-Roses, France 5 Université Paris-Diderot, 75013 Paris 7, France 6 Université Paris-Saclay, 78140 Paris 11, France * Correspondence: [email protected] (V.H.); [email protected] (N.O.F.); Tel.: +33-1-48-68-96-00 (V.H.); +33-1-60-87-34-92 or +33-1-60-87-34-98 (N.O.F.) These authors contributed equally to the work. y Received: 15 December 2019; Accepted: 24 January 2020; Published: 5 February 2020 Abstract: Human skin dermis contains fibroblast subpopulations in which characterization is crucial due to their roles in extracellular matrix (ECM) biology. -

Canonical TGF Signaling and Its Contribution to Endometrial Cancer
Journal of Clinical Medicine Review Canonical TGFβ Signaling and Its Contribution to Endometrial Cancer Development and Progression—Underestimated Target of Anticancer Strategies Piotr K. Zakrzewski Department of Cytobiochemistry, Faculty of Biology and Environmental Protection, University of Lodz, Pomorska 141/143, 90-236 Lodz, Poland; [email protected]; Tel.: +48-42-635-52-99 Abstract: Endometrial cancer is one of the leading gynecological cancers diagnosed among women in their menopausal and postmenopausal age. Despite the progress in molecular biology and medicine, no efficient and powerful diagnostic and prognostic marker is dedicated to endometrial carcinogenesis. The canonical TGFβ pathway is a pleiotropic signaling cascade orchestrating a variety of cellular and molecular processes, whose alterations are responsible for carcinogenesis that originates from different tissue types. This review covers the current knowledge concerning the canonical TGFβ pathway (Smad-dependent) induced by prototypical TGFβ isoforms and the involvement of pathway alterations in the development and progression of endometrial neoplastic lesions. Since Smad-dependent signalization governs opposed cellular processes, such as growth arrest, apoptosis, tumor cells growth and differentiation, as well as angiogenesis and metastasis, TGFβ cascade may act both as a tumor suppressor or tumor promoter. However, the final effect of TGFβ signaling on endometrial cancer cells depends on the cancer disease stage. The multifunctional Citation: Zakrzewski, P.K. Canonical role of the TGFβ pathway indicates the possible utilization of alterations in the TGFβ cascade as a TGFβ Signaling and Its Contribution potential target of novel anticancer strategies. to Endometrial Cancer Development and Progression—Underestimated Keywords: endometrial cancer; TGFβ isoforms; TGFβR1; TGFβR2; Smad proteins; TGFβ co-receptors; Target of Anticancer Strategies. -

MOLECULAR GENETI a Thesis Submitted to the School Of
MOLECULAR GENETICS OF CATTLE MUSCULARITY By Irida Novianti A thesis submitted to the University of Adelaide in fulfilment of the requirement of the degree of Master of Agricultural Science The University of Adelaide School of Animal and Veterinary Science December 2010 DECLARATION I declare that this thesis is a record of original work and contains no material that has been accepted for the award of any other degree or diploma in any university or other tertiary institution to Irida Novianti. To the best of my knowledge and belief, this thesis contains no material previously published or written by any other person, except where due reference is made in the text. I give consent to this copy of my thesis, when deposited in the University Library, being made available for loan and photocopying, subject to the provisions of the Copyright Act 1968. I also give permission for the digital version of my thesis to be made available on the web, via the University’s digital research repository, the Library catalogue, the Australasian Digital Theses Program (ADTP) and also through web search engines, unless permission has been granted by the University to restrict access for a period of time. Irida Novianti December, 2010 ii TABLE OF CONTENTS Declaration ........................................................................................................... ii Index of Figures ................................................................................................... vi Index of Tables .................................................................................................... -

Snail Regulates BMP and Tgfβ Pathways to Control the Differentiation Status of Glioma-Initiating Cells
Oncogene (2018) 37:2515–2531 https://doi.org/10.1038/s41388-018-0136-0 ARTICLE Snail regulates BMP and TGFβ pathways to control the differentiation status of glioma-initiating cells 1,2 1,2 1,2 3 1,2 1,2,3 Laia Caja ● Kalliopi Tzavlaki ● Mahsa S. Dadras ● E-Jean Tan ● Gad Hatem ● Naga P. Maturi ● 1,2 4 2,5 2,6 4 3 Anita Morén ● Lotta Wik ● Yukihide Watanabe ● Katia Savary ● Masood Kamali-Moghaddan ● Lene Uhrbom ● 1,2 1,2 Carl-Henrik Heldin ● Aristidis Moustakas Received: 14 June 2017 / Revised: 12 December 2017 / Accepted: 28 December 2017 / Published online: 16 February 2018 © The Author(s) 2018. This article is published with open access Abstract Glioblastoma multiforme is a brain malignancy characterized by high heterogeneity, invasiveness, and resistance to current therapies, attributes related to the occurrence of glioma stem cells (GSCs). Transforming growth factor β (TGFβ) promotes self-renewal and bone morphogenetic protein (BMP) induces differentiation of GSCs. BMP7 induces the transcription factor Snail to promote astrocytic differentiation in GSCs and suppress tumor growth in vivo. We demonstrate that Snail represses stemness in GSCs. Snail interacts with SMAD signaling mediators, generates a positive feedback loop of BMP signaling and β β 1234567890();,: transcriptionally represses the TGFB1 gene, decreasing TGF 1 signaling activity. Exogenous TGF 1 counteracts Snail function in vitro, and in vivo promotes proliferation and re-expression of Nestin, confirming the importance of TGFB1 gene repression by Snail. In conclusion, novel insight highlights mechanisms whereby Snail differentially regulates the activity of the opposing BMP and TGFβ pathways, thus promoting an astrocytic fate switch and repressing stemness in GSCs. -

Mouse TGFBR3 / Betaglycan Protein (His Tag)
Mouse TGFBR3 / Betaglycan Protein (His Tag) Catalog Number: 50542-M08H General Information SDS-PAGE: Gene Name Synonym: 1110036H20Rik; AU015626; AW215636; TBRIII Protein Construction: A DNA sequence encoding the extracellular domain of mouse TGFBR3 (NP_072075.2) (Met 1-Arg 399) was expressed, with a polyhistidine tag at the C-terminus. Source: Mouse Expression Host: HEK293 Cells QC Testing Purity: > 95 % as determined by SDS-PAGE Endotoxin: Protein Description < 1.0 EU per μg of the protein as determined by the LAL method Betaglycan also known as transforming growth factor beta receptor III (TGFBR3), is a cell-surface chondroitin sulfate / heparan sulfate Stability: proteoglycan. TGFBR3 is a transforming growth factor (TGF)-beta type III receptor. This receptor is a membrane proteoglycan that often functions as Samples are stable for up to twelve months from date of receipt at -70 ℃ a co-receptor with other TGF-beta receptor superfamily members. Ectodomain shedding produces soluble TGFBR3, which may inhibit TGFB Predicted N terminal: Gly 23 signaling. Decreased expression of this receptor has been observed in Molecular Mass: various cancers. TGFBR3 is the TGF-β component most commonly downregulated among localized human prostate cancer studies. TGFBR3 The recombinant mouse TGFBR3 consists of 774 amino acids and has a knockdown led to focus formation and enhanced expression of CD133, a predicted molecular mass of 86.3 kDa. In SDS-PAGE under reducing marker found on prostate cancer stem cells. TGFBR3 is an accessory conditions, the apparent molecular mass of rmTGFBR3 is approximately receptor that binds to and modulates the activities of both transforming 80-90 kDa due to glycosylation. -

Glucocorticoids Recruit Tgfbr3 and Smad1 to Shift Transforming Growth Factor-Β Signaling from the Tgfbr1/Smad2/3 Axis to the Acvrl1/Smad1 Axis in Lung Fibroblasts
Glucocorticoids recruit Tgfbr3 and Smad1 to shift transforming growth factor-β signaling from the Tgfbr1/Smad2/3 axis to the Acvrl1/Smad1 axis in lung fibroblasts Inauguraldissertation zur Erlangung des Grades eines Doktors der Medizin des Fachbereichs Medizin der Justus-Liebig-Universität Gießen vorgelegt von Schwartze, Julian Tristan aus Langen (Hessen), Deutschland Gießen (2014) Aus dem Department of Lung Development and Remodelling, Max Planck Institute for Heart and Lung Research, Bad Nauheim, Germany; Department of Internal Medicine (Pulmonology), University of Giessen and Marburg Lung Center, German Center for Lung Research, Giessen, Germany. Leiter/Direktor: Prof. Dr. med. Werner Seeger Gutachter: Prof. Dr. med. Werner Seeger Gutachter: Prof. Dr. Klaus T. Preissner Tag der Disputation: 31.07.2015 Table of contents I I. Table of contents I. Table of contents ................................................................................................ I II. List of figures .................................................................................................... VI III. List of tables ................................................................................................... VIII IV. List of abbreviations ......................................................................................... IX V. Summary ........................................................................................................... XI VI. Zusammenfassung ......................................................................................... -
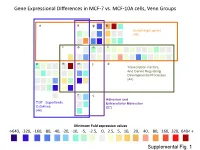
Supplementary Data
Gene Expressional Differences in MCF-7 vs. MCF-10A cells, Venn Groups e k g b Smad target genes (48) l o n i a h m j d Transcription Factors, And Genes Regulating Developmental Processes (44) f c Adhesion and TGF Superfamily Extracellular Molecules Cytokines (27) (44) Minimum Fold expression values ->640, -320, -160, -80, -40, -20, -10, -5, -2.5, 0, 2.5, 5, 10, 20, 40, 80, 160, 320, 640< + Supplemental Fig. 1 Supplemental Table 1. Change of Basal Gene Expressional values in MCF-7 as compared to MCF-10A cell line. Fold T-test Gene # GeneBank Symbol Up/Down p-Value Description /Position Regulation Venn Group a 01 /A01 NM_001105 ACVR1 -4.96 0.0000 Activin A receptor, type I 02 /A02 NM_001616 ACVR2A -2.62 0.0007 Activin A receptor, type IIA 03 /A03 NM_000020 ACVRL1 -1.25 0.7013 Activin A receptor type II-like 1 05 /A05 NM_020547 AMHR2 1.08 0.8043 Anti-Mullerian hormone receptor, type II 16 /B04 NM_004329 BMPR1A -2.40 0.0011 Bone morphogenetic protein receptor, type IA 17 /B05 NM_001203 BMPR1B -7.37 0.0000 Bone morphogenetic protein receptor, type IB 36 /C12 NM_000557 GDF5 -1.38 0.4911 Growth differentiation factor 5 (cartilage-derived morphogenetic protein-1) 37 /D01 NM_001001557 GDF6 1.12 0.9002 Growth differentiation factor 6 38 /D02 NM_182828 GDF7 -1.52 0.4995 Growth differentiation factor 7 53 /E05 NM_020997 LEFTY1 -1.76 0.0529 Left-right determination factor 1 59 /E11 NM_018055 NODAL -3.62 0.1290 Nodal homolog (mouse) 77 /G05 NM_003238 TGFB2 -4.52 0.0566 Transforming growth factor, beta 2 78 /G06 NM_003239 TGFB3 -1.12 0.2902 Transforming -

Gastrocnemius Transcriptome Analysis Reveals Domestication Induced Gene Expression Changes Between Wild and Domestic Chickens
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector Genomics 100 (2012) 314–319 Contents lists available at SciVerse ScienceDirect Genomics journal homepage: www.elsevier.com/locate/ygeno Gastrocnemius transcriptome analysis reveals domestication induced gene expression changes between wild and domestic chickens Qinghe Li a,1, Nan Wang b,1, Zhuo Du a, Xiaoxiang Hu a, Li Chen c, Jing Fei a, Yuanyuan Wang a, Ning Li a,⁎ a State Key Laboratory for Agro-biotechnology, China Agricultural University, Beijing, 100193, China b College of Veterinary Medicine, China Agricultural University, Beijing 100193, China c Institute of Animal Sciences and Veterinary Medicine, Zhejiang Academy of Agricultural Sciences, Hangzhou, 310021, China article info abstract Article history: Artificial selection of chicken for human-preferred traits has manifested great phenotypic differences be- Received 19 January 2012 tween wild and domestic chickens. Study on the formation of these phenotypic variations will contribute Accepted 12 July 2012 to comprehensive understanding of the molecular mechanism of animal domestication. We used three Available online 21 July 2012 kinds of chicken breeds for transcriptome analysis, including the red jungle fowl which was the wild ancestor of chickens, and two other domestic breeds, the chahua chicken and the avian broiler. More than 12,000 Keywords: genes' expression levels were compared between different chicken breeds, and hundreds of genes displayed Wild chicken Domestic chicken differential expression levels compared with wild chicken. Gene ontology analysis showed that differentially Gene expression profile expressed genes in domestic chickens tended to be enriched in extracellular matrix, DNA binding and im- Domestication mune system development, etc. -

MOLECULAR GENETI a Thesis Submitted to the School Of
MOLECULAR GENETICS OF CATTLE MUSCULARITY By Irida Novianti A thesis submitted to the University of Adelaide in fulfilment of the requirement of the degree of Master of Agricultural Science The University of Adelaide School of Animal and Veterinary Science December 2010 DECLARATION I declare that this thesis is a record of original work and contains no material that has been accepted for the award of any other degree or diploma in any university or other tertiary institution to Irida Novianti. To the best of my knowledge and belief, this thesis contains no material previously published or written by any other person, except where due reference is made in the text. I give consent to this copy of my thesis, when deposited in the University Library, being made available for loan and photocopying, subject to the provisions of the Copyright Act 1968. I also give permission for the digital version of my thesis to be made available on the web, via the University’s digital research repository, the Library catalogue, the Australasian Digital Theses Program (ADTP) and also through web search engines, unless permission has been granted by the University to restrict access for a period of time. Irida Novianti December, 2010 ii TABLE OF CONTENTS Declaration ........................................................................................................... ii Index of Figures ................................................................................................... vi Index of Tables ....................................................................................................