Identication of Key Player Genes in Gene Regulatory Networks
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Amplification of 8Q21 in Breast Cancer Is Independent of MYC and Associated with Poor Patient Outcome
Modern Pathology (2010) 23, 603–610 & 2010 USCAP, Inc. All rights reserved 0893-3952/10 $32.00 603 Amplification of 8q21 in breast cancer is independent of MYC and associated with poor patient outcome Matthias Choschzick1, Paula Lassen1, Annette Lebeau1, Andreas Holger Marx1, Luigi Terracciano2, Uwe Heilenko¨tter3, Fritz Jaenicke4, Carsten Bokemeyer5, Jakob Izbicki6, Guido Sauter1 and Ronald Simon1 1Institute of Pathology, University Medical Centre Hamburg-Eppendorf, Hamburg, Germany; 2Institute of Pathology, University Hospital Basel, Basel, Switzerland; 3Department of Gynaecology, Hospital Itzehoe, Itzehoe, Germany; 4Department of Gynaecology, University Medical Centre Hamburg-Eppendorf, Hamburg, Germany; 5Department of Oncology, Hematology, Bone Marrow Transplantation with Section Pneumology, University Hospital Hamburg-Eppendorf, Hamburg, Germany and 6Department of Surgery, University Medical Centre Hamburg-Eppendorf, Hamburg, Germany Copy number gains involving the long arm of chromosome 8, including high-level amplifications at 8q21 and 8q24, have been frequently reported in breast cancer. Although the role of the MYC gene as the driver of the 8q24 amplicon is well established, the significance of the 8q21 amplicon is less clear. The breast cancer cell line SK-BR-3 contains three separate 8q21 amplicons, the distal two of which correspond to putative target genes TPD52 and WWP1. To understand the effect of proximal 8q21 amplification on breast cancer phenotype and patient prognosis, we analyzed 8q21 copy number changes using fluorescence in situ hybridization (FISH) in a tissue microarray containing more than 2000 breast cancers. Amplification at 8q21 was found in 3% of tumors, and was associated with medullary type (Po0.03), high tumor grade (Po0.0001), high Ki67 labeling index (Po0.05), amplification of MYC (Po0.0001), HER2, MDM2, and CCND1 (Po0.05 each), as well as the total number of gene amplifications (Po0.0001). -

Differential Expression Profiling of Gene Response to Ionizing Radiation in Two Endometrial Cancer Cell Lines with Distinct Radiosensitivities
625-634 28/1/2009 12:32 ÌÌ ™ÂÏ›‰·625 ONCOLOGY REPORTS 21: 625-634, 2009 625 Differential expression profiling of gene response to ionizing radiation in two endometrial cancer cell lines with distinct radiosensitivities XUE-LIAN DU1,2, TAO JIANG2, ZE-QING WEN1, QING-SHUI LI2, RONG GAO2 and FEI WANG1 1Department of Gynecologic Oncology, Shandong Tumor Hospital, Shandong University, Jinan 250117; 2Department of Obstetrics and Gynecology, Provincial Hospital Affiliated to Shandong University, Jinan 250021, P.R. China Received November 4, 2008; Accepted December 17, 2008 DOI: 10.3892/or_00000265 Abstracts. Although radiotherapy is routinely administered Introduction to high-risk endometrial carcinoma and offer a significant disease-free survival advantage, the therapeutic effect is Endometrial cancer is one of the most common gynecological sometimes limited by the occurrence of radioresistance. To malignancies worldwide. Surgery is the preferred initial determine the patterns of gene expression responsible for treatment and most women with early-stage, low-risk disease the radioresistance and to search for potential target genes for will do well without adjuvant radiotherapy. However, both radiotherapy, we selected two cell lines with distinct radio- intermediate-risk and high-risk patients are at risk for local- sensitivities using colony-formation assay from four endo- regional relapse and therefore adjuvant radiotherapy, such as metrial cancer cell lines. The cell cycle distribution showed pelvic radiation, vaginal brachytherapy, and whole-abdomen higher fractions of G2/M phase cells in the radiosensitive cell radiation, is essential for local control (1). Johnson and line KLE after radiation compared with the radioresistant cell colleagues reported that adjuvant external-beam pelvic radio- line ISK. -

Human Diseases Associated with Defects in Assembly of OXPHOS Complexes
Essays in Biochemistry (2018) 62 271–286 https://doi.org/10.1042/EBC20170099 Review Article Human diseases associated with defects in assembly of OXPHOS complexes Daniele Ghezzi1,2 and Massimo Zeviani3 1Molecular Neurogenetics, Foundation IRCCS Neurological Institute Besta, Milan, Italy; 2Department of Pathophysiology and Transplantation, University of Milan, Milan, Italy; 3Medical Research Council – Mitochondrial Biology Unit, University of Cambridge, Cambridge, U.K. Correspondence: Massimo Zeviani ([email protected]) The structural biogenesis and functional proficiency of the multiheteromeric complexes forming the mitochondrial oxidative phosphorylation system (OXPHOS) require the con- certed action of a number of chaperones and other assembly factors, most of which are specific for each complex. Mutations in a large number of these assembly factors arere- sponsible for mitochondrial disorders, in most cases of infantile onset, typically character- ized by biochemical defects of single specific complexes. In fact, pathogenic mutations in complex-specific assembly factors outnumber, in many cases, the repertoire of mutations found in structural subunits of specific complexes. The identification of patients with spe- cific defects in assembly factors has provided an important contribution to the nosological characterization of mitochondrial disorders, and has also been a crucial means to identify a huge number of these proteins in humans, which play an essential role in mitochondrial bioenergetics. The wide use of next generation sequencing (NGS) has led to and will allow the identifcation of additional components of the assembly machinery of individual com- plexes, mutations of which are responsible for human disorders. The functional studies on patients’ specimens, together with the creation and characterization of in vivo models, are fundamental to better understand the mechanisms of each of them. -

Porta-SIMD: an Optimally Portable SIMD Programming Language Duke CS-1990-12 UNC CS TR90-021 May 1990
Porta-SIMD: An Optimally Portable SIMD Programming Language Duke CS-1990-12 UNC CS TR90-021 May 1990 Russ Tuck Duke University Deparment of Computer Science Durham, NC 27706 The University of North Carolina at Chapel Hill Department of Computer Science CB#3175, Sitterson Hall Chapel Hill, NC 27599-3175 Text (without appendix) of a Ph.D. dissertation submitted to Duke University. The research was performed at UNC. @ 1990 Russell R. Tuck, III UNC is an Equal Opportunity/Atlirmative Action Institution. PORTA-SIMD: AN OPTIMALLY PORTABLE SIMD PROGRAMMING LANGUAGE by Russell Raymond Tuck, III Department of Computer Science Duke University Dissertation submitte in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Department of Computer Science in the Graduate School of Duke University 1990 Copyright © 1990 by Russell Raymond Tuck, III All rights reserved Abstract Existing programming languages contain architectural assumptions which limit their porta bility. I submit optimal portability, a new concept which solves this language design problem. Optimal portability makes it possible to design languages which are portable across vari ous sets of diverse architectures. SIMD (Single-Instruction stream, Multiple-Data stream) computers represent an important and very diverse set of architectures for which to demon strate optimal portability. Porta-SIMD (pronounced "porta.-simm'd") is the first optimally portable language for SIMD computers. It was designed and implemented to demonstrate that optimal portability is a useful and achievable standard for language design. An optimally portable language allows each program to specify the architectural features it requires. The language then enables the compiled program to exploit exactly those fea. tures, and to run on all architectures that provide them. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Supporting Information
Supporting Information Edgar et al. 10.1073/pnas.1601895113 SI Methods (Actimetrics), and recordings were analyzed using LumiCycle Mice. Sample size was determined using the resource equation: Data Analysis software (Actimetrics). E (degrees of freedom in ANOVA) = (total number of exper- – Cell Cycle Analysis of Confluent Cell Monolayers. NIH 3T3, primary imental animals) (number of experimental groups), with −/− sample size adhering to the condition 10 < E < 20. For com- WT, and Bmal1 fibroblasts were sequentially transduced − − parison of MuHV-4 and HSV-1 infection in WT vs. Bmal1 / with lentiviral fluorescent ubiquitin-based cell cycle indicators mice at ZT7 (Fig. 2), the investigator did not know the genotype (FUCCI) mCherry::Cdt1 and amCyan::Geminin reporters (32). of the animals when conducting infections, bioluminescence Dual reporter-positive cells were selected by FACS (Influx Cell imaging, and quantification. For bioluminescence imaging, Sorter; BD Biosciences) and seeded onto 35-mm dishes for mice were injected intraperitoneally with endotoxin-free lucif- subsequent analysis. To confirm that expression of mCherry:: Cdt1 and amCyan::Geminin correspond to G1 (2n DNA con- erin (Promega E6552) using 2 mg total per mouse. Following < ≤ anesthesia with isofluorane, they were scanned with an IVIS tent) and S/G2 (2 n 4 DNA content) cell cycle phases, Lumina (Caliper Life Sciences), 15 min after luciferin admin- respectively, cells were stained with DNA dye DRAQ5 (abcam) and analyzed by flow cytometry (LSR-Fortessa; BD Biosci- istration. Signal intensity was quantified using Living Image ences). To examine dynamics of replicative activity under ex- software (Caliper Life Sciences), obtaining maximum radiance perimental confluent conditions, synchronized FUCCI reporter for designated regions of interest (photons per second per − − − monolayers were observed by time-lapse live cell imaging over square centimeter per Steradian: photons·s 1·cm 2·sr 1), relative 3 d (Nikon Eclipse Ti-E inverted epifluorescent microscope). -

Roles of TBC1D1 and TBC1D4 in Insulin- and Exercise-Stimulated Glucose Transport of Skeletal Muscle
Diabetologia (2015) 58:19–30 DOI 10.1007/s00125-014-3395-5 REVIEW Roles of TBC1D1 and TBC1D4 in insulin- and exercise-stimulated glucose transport of skeletal muscle Gregory D. Cartee Received: 30 June 2014 /Accepted: 7 August 2014 /Published online: 4 October 2014 # Springer-Verlag Berlin Heidelberg 2014 Abstract This review focuses on two paralogue Rab GTPase mechanism for greater TBC1D4 phosphorylation in insulin- activating proteins known as TBC1D1 Tre-2/BUB2/cdc 1 stimulated muscles after acute exercise is uncertain, and a domain family (TBC1D) 1 and TBC1D4 (also called Akt causal link between enhanced TBC1D4 phosphorylation and Substrate of 160 kDa, AS160) and their roles in controlling increased post-exercise insulin sensitivity has yet to be skeletal muscle glucose transport in response to the indepen- established. In summary, TBC1D1 and TBC1D4 have impor- dent and combined effects of insulin and exercise. Convincing tant, but distinct roles in regulating muscle glucose transport evidence implicates Akt2-dependent TBC1D4 phosphoryla- in response to insulin and exercise. tion on T642 as a key part of the mechanism for insulin- stimulated glucose uptake by skeletal muscle. TBC1D1 phos- Keywords Akt substrate of 160 kDa . Diabetes . Glucose phorylation on several insulin-responsive sites (including transport .High-fatdiet .Insulinresistance .Obesity .Physical T596, a site corresponding to T642 in TBC1D4) does not activity . Review appear to be essential for in vivo insulin-stimulated glucose uptake by skeletal muscle. In vivo exercise or ex vivo con- Abbreviations traction of muscle result in greater TBC1D1 phosphorylation AMPK 5' AMP-activated kinase on S237 that is likely to be secondary to increased AMP- APPL2 Adaptor protein containing PH domain, activated protein kinase activity and potentially important for PTB domain and leucine zipper motif 2 contraction-stimulated glucose uptake. -

Pnw 2020 Strunk001.Pdf
Remote Sensing of Environment 237 (2020) 111535 Contents lists available at ScienceDirect Remote Sensing of Environment journal homepage: www.elsevier.com/locate/rse Evaluation of pushbroom DAP relative to frame camera DAP and lidar for forest modeling T ∗ Jacob L. Strunka, , Peter J. Gouldb, Petteri Packalenc, Demetrios Gatziolisd, Danuta Greblowskae, Caleb Makif, Robert J. McGaugheyg a USDA Forest Service Pacific Northwest Research Station, 3625 93rd Ave SW, Olympia, WA, 98512, USA b Washington State Department of Natural Resources, PO Box 47000, 1111 Washington Street, SE, Olympia, WA, 98504-7000, USA c School of Forest Sciences, Faculty of Science and Forestry, University of Eastern Finland, P.O. Box 111, 80101, Joensuu, Finland d USDA Forest Service Pacific Northwest Research Station, 620 Southwest Main, Suite 502, Portland, OR, 97205, USA e GeoTerra Inc., 60 McKinley St, Eugene, OR, 97402, USA f Washington State Department of Natural Resources, PO Box 47000, 1111 Washington Street SE, Olympia, WA, 98504-7000, USA g USDA Forest Service Pacific Northwest Research Station, University of Washington, PO Box 352100, Seattle, WA, 98195-2100, USA ARTICLE INFO ABSTRACT Keywords: There is growing interest in using Digital Aerial Photogrammetry (DAP) for forestry applications. However, the Lidar performance of pushbroom DAP relative to frame-based DAP and airborne lidar is not well documented. Interest Structure from motion in DAP stems largely from its low cost relative to lidar. Studies have demonstrated that frame-based DAP Photogrammetry generally performs slightly poorer than lidar, but still provides good value due to its reduced cost. In the USA Forestry pushbroom imagery can be dramatically less expensive than frame-camera imagery in part because of a na- DAP tionwide collection program. -

Aneuploidy: Using Genetic Instability to Preserve a Haploid Genome?
Health Science Campus FINAL APPROVAL OF DISSERTATION Doctor of Philosophy in Biomedical Science (Cancer Biology) Aneuploidy: Using genetic instability to preserve a haploid genome? Submitted by: Ramona Ramdath In partial fulfillment of the requirements for the degree of Doctor of Philosophy in Biomedical Science Examination Committee Signature/Date Major Advisor: David Allison, M.D., Ph.D. Academic James Trempe, Ph.D. Advisory Committee: David Giovanucci, Ph.D. Randall Ruch, Ph.D. Ronald Mellgren, Ph.D. Senior Associate Dean College of Graduate Studies Michael S. Bisesi, Ph.D. Date of Defense: April 10, 2009 Aneuploidy: Using genetic instability to preserve a haploid genome? Ramona Ramdath University of Toledo, Health Science Campus 2009 Dedication I dedicate this dissertation to my grandfather who died of lung cancer two years ago, but who always instilled in us the value and importance of education. And to my mom and sister, both of whom have been pillars of support and stimulating conversations. To my sister, Rehanna, especially- I hope this inspires you to achieve all that you want to in life, academically and otherwise. ii Acknowledgements As we go through these academic journeys, there are so many along the way that make an impact not only on our work, but on our lives as well, and I would like to say a heartfelt thank you to all of those people: My Committee members- Dr. James Trempe, Dr. David Giovanucchi, Dr. Ronald Mellgren and Dr. Randall Ruch for their guidance, suggestions, support and confidence in me. My major advisor- Dr. David Allison, for his constructive criticism and positive reinforcement. -
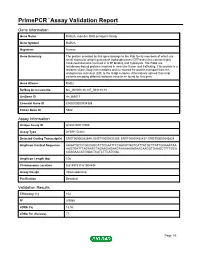
Download Validation Data
PrimePCR™Assay Validation Report Gene Information Gene Name RAB2A, member RAS oncogene family Gene Symbol RAB2A Organism Human Gene Summary The protein encoded by this gene belongs to the Rab family members of which are small molecular weight guanosine triphosphatases (GTPases) that contain highly conserved domains involved in GTP binding and hydrolysis. The Rabs are membrane-bound proteins involved in vesicular fusion and trafficking. This protein is a resident of pre-Golgi intermediates and is required for protein transport from the endoplasmic reticulum (ER) to the Golgi complex. Alternatively spliced transcript variants encoding different isoforms have been found for this gene. Gene Aliases RAB2 RefSeq Accession No. NC_000008.10, NT_008183.19 UniGene ID Hs.369017 Ensembl Gene ID ENSG00000104388 Entrez Gene ID 5862 Assay Information Unique Assay ID qHsaCID0011980 Assay Type SYBR® Green Detected Coding Transcript(s) ENST00000262646, ENST00000531289, ENST00000452437, ENST00000543829 Amplicon Context Sequence AAGATGCCCGCCAGCATTCCAATTCCAACATGGTCATTATGCTTATTGGAAATAA AAGTGATTTAGAATCTAGAAGAGAAGTAAAAAAAGAAGAAGGTGAAGCTTTTGCA CGAGAACATGGACTCATCTTCATGGA Amplicon Length (bp) 106 Chromosome Location 8:61497317-61504494 Assay Design Intron-spanning Purification Desalted Validation Results Efficiency (%) 104 R2 0.9988 cDNA Cq 18.53 cDNA Tm (Celsius) 77 Page 1/5 PrimePCR™Assay Validation Report gDNA Cq 43.2 Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 2/5 PrimePCR™Assay Validation Report -

GUIDE to INTERNATIONAL UNIVERSITY ADMISSION About NACAC
GUIDE TO INTERNATIONAL UNIVERSITY ADMISSION About NACAC The National Association for College Admission Counseling (NACAC), founded in 1937, is an organization of 14,000 professionals from around the world dedicated to serving students as they make choices about pursuing postsecondary education. NACAC is committed to maintaining high standards that foster ethical and social responsibility among those involved in the transition process, as outlined in the NACAC’s Guide to Ethical Practice in College Admission. For more information and resources, visit nacacnet.org. The information presented in this document may be reprinted and distributed with permission from and attribution to the National Association for College Admission Counseling. It is intended as a general guide and is presented as is and without warranty of any kind. While every effort has been made to ensure the accuracy of the content, NACAC shall not in any event be liable to any user or any third party for any direct or indirect loss or damage caused or alleged to be caused by the information contained herein and referenced. Copyright © 2020 by the National Association for College Admission Counseling. All rights reserved. NACAC 1050 N. Highland Street Suite 400 Arlington, VA 22201 800.822.6285 nacacnet.org COVID-19 IMPACTS ON APPLYING ABROAD NACAC is pleased to offer this resource for the fifth year. NACAC’s Guide to International University Admission promotes study options outside students’ home countries for those who seek an international experience. Though the impact the current global health crisis will have on future classes remains unclear, we anticipate that there will still be a desire among students—perhaps enhanced as a result of COVID-19, to connect with people from other cultures and parts of the world, and to pursue an undergraduate degree abroad. -

Mitochondrial Medicine in the Omics Era
Mitochondrial Medicine in the Omics Era Joyeeta Rahman1 and Shamima Rahman1,2* 1 Mitochondrial Research Group, UCL Great Ormond Street Institute of Child Health and 2 Metabolic Unit, Great Ormond Street Hospital NHS Foundation Trust, London, UK *Correspondence to: Professor Shamima Rahman Mitochondrial Research Group Genetics and Genomic Medicine UCL Great Ormond Street Institute of Child Health London WC1N 1EH, UK. Telephone: +44 (0)2079052608 [email protected] Keywords: Mitochondrial disease, OXPHOS, signalling, omics, genomics, transcriptomics, proteomics, metabolomics, mitochondrial stress response, treatment 1 Abstract Mitochondria are dynamic bioenergetic organelles whose maintenance requires ~1500 proteins from two genomes. Mutations in either the mitochondrial or nuclear genome can disrupt a plethora of cellular metabolic and homeostatic functions. Mitochondrial diseases represent one the most common and severe groups of inherited genetic disorders, characterised by clinical, biochemical, and genetic heterogeneity, diagnostic odysseys, and lack of curative therapies. This review aims to discuss recent advances in mitochondrial biology and medicine arising from widespread utilisation of high-throughput omics technologies, and also includes a broad discussion of emerging therapies for mitochondrial disease. New insights into both bioenergetic and biosynthetic mitochondrial functionalities have expedited the genetic diagnosis of primary mitochondrial disorders, and identified novel mitochondrial pathomechanisms and new targets