Supplemental Material 1 (PDF)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
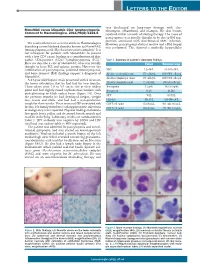
Letters to the Editor
LETTERS TO THE EDITOR was discharged on long-term therapy with clar - + MonoMAC versus idiopathic CD4 lymphocytopenia . ithromycin, ethambutol, and rifampin. His skin lesions Comment to Haematologica. 2011;96(8):1221-5 resolved within a month of starting therapy. The cause of pancytopenia was initially thought to be due to BM sup - pression associated with disseminated MAC infection. We read with interest a recent article in Haematologica However, pancytopenia did not resolve and a BM biopsy describing a novel defined disorder known as MonoMAC was performed. This showed a markedly hypocellular (monocytopenia with Mycobacterium avium complex). 1 It is not infrequent for patients with MonoMAC to present with a low CD4 count, leading to a consideration of idio - pathic CD4-positive (CD4 +) lymphocytopenia (ICL). 2 Table 1. Summary of patient’s laboratory findings. Here we describe a case of MonoMAC who was initially Patient Reference range thought to have ICL and aplastic anemia. However, the combination of pancytopenia, profound monocytopenia, WBC 1.2 ¥10 9/L 4.1-10.9 ¥10 9/L and bone marrow (BM) findings support a diagnosis of Absolute neutrophil count 876 cells/ mL 1400-6500 cells/ mL MonoMAC. Absolute lymphocyte count 144 cells/ mL 1200-3400 cells/ mL A 24-year old Hispanic male presented with 2 ulcers on the lower extremities that he had had for two months. Absolute monocyte count 12 cells/ mL 300-820 cells/ mL These ulcers were 1.0 to 3.5 cm in size at their widest Hemoglobin 5.2 g/dL 14.0-18.0 g/dL point and had slightly raised erythematous borders with Hematocrit 16.2% 40-54% pink-glistening to black eschar bases (Figure 1A). -

Editorials & Perspectives
EDITORIALS & PERSPECTIVES Dendritic cell, monocyte, B and NK lymphoid deficiency defines the lost lineages of a new GATA-2 dependent myelodysplastic syndrome Venetia Bigley and Matthew Collin Institute of Cellular Medicine, Newcastle University, UK. E-mail: [email protected] doi:10.3324/haematol.2011.048355 (Related Original Article on page 1221) novel immunodeficiency syndrome has recently been International Prognostic Scoring System (IPSS). defined in young adults. Known variously as dendritic It is worth emphasizing at this point that DCML deficiency cell, monocyte, B and NK lymphoid (DCML deficien - patients may have significant risk to their health before addi - A1 2 2 cy), ‘autosomal dominant and sporadic monocytopenia’ or tional cytopenias evolve. Disseminated mycobacterial infec - ‘MonoMAC’ (monocytopenia with Mycobacterium avium com - tion, PAP and carcinoma in situ may all occur prior to the onset plex), 3 it is characterized by a composite mononuclear cell defi - of an obvious myelodysplastic syndrome, underscoring the ciency, atypical mycobacterial and viral infection, and progres - fact that immunodeficiency is a cardinal feature of the disor - sion to myelodysplasia and leukemia. Strikingly, a number of der. In the series of 4 subjects described in 2011, one had mild patients also develop pulmonary alveolar proteinosis (PAP). anemia (10.3 g/dL) and one mild thrombocytopenia Patients typically present in their 3 rd or 4 th decade, although his - (123 ¥10 9/L), yet 2 required hematopoietic stem cell transplan - torical blood counts may show pre-existing monocytopenia tation (one for refractory mycobacterial infection, one for PAP) for ten years or more. A substantial number of patients devel - and a third died of influenza H1N1 prior to transplantation. -

PBX3 and MEIS1 Cooperate in Hematopoietic Cells to Drive Acute
Published OnlineFirst January 8, 2016; DOI: 10.1158/0008-5472.CAN-15-1566 Cancer Molecular and Cellular Pathobiology Research PBX3 and MEIS1 Cooperate in Hematopoietic Cells to Drive Acute Myeloid Leukemias Characterized by a Core Transcriptome of the MLL-Rearranged Disease Zejuan Li1, Ping Chen1, Rui Su2, Chao Hu1,2,3, Yuanyuan Li1, Abdel G. Elkahloun4, Zhixiang Zuo1,2, Sandeep Gurbuxani5, Stephen Arnovitz1, Hengyou Weng1,2, Yungui Wang1,2,3, Shenglai Li1, Hao Huang1, Mary Beth Neilly1, Gang Greg Wang6, Xi Jiang1,2, Paul P. Liu4, Jie Jin3, and Jianjun Chen1,2 Abstract Overexpression of HOXA/MEIS1/PBX3 homeobox genes is sion profiling of hematopoietic cells demonstrated that PBX3/ the hallmark of mixed lineage leukemia (MLL)-rearranged MEIS1 overexpression, but not HOXA9/MEIS1, HOXA9/PBX3, acute myeloid leukemia (AML). HOXA9 and MEIS1 are con- or HOXA9 overexpression, recapitulated the MLL-fusion–medi- sideredtobethemostcriticaltargetsofMLLfusionsandtheir ated core transcriptome, particularly upregulation of the endog- coexpression rapidly induces AML. MEIS1 and PBX3 are not enous Hoxa genes. Disruption of the binding between MEIS1 individually able to transform cells and were therefore hypoth- andPBX3diminishedPBX3/MEIS1–mediated cell transforma- esized to function as cofactors of HOXA9. However, in this tion and HOX gene upregulation. Collectively, our studies study, we demonstrate that coexpression of PBX3 and MEIS1 strongly implicate the PBX3/MEIS1 interaction as a driver of (PBX3/MEIS1), without ectopic expression of a HOX gene, is cell transformation and leukemogenesis, and suggest that this sufficient for transformation of normal mouse hematopoietic axis may play a critical role in the regulation of the core stem/progenitor cells in vitro.Moreover,PBX3/MEIS1 overex- transcriptional programs activated in MLL-rearranged and pression also caused AML in vivo, with a leukemic latency HOX-overexpressing AML. -

Secondary Pulmonary Alveolar Proteinosis in Hematologic
review Secondary pulmonary alveolar proteinosis in hematologic malignancies Chakra P Chaulagain a,*, Monika Pilichowska b, Laurence Brinckerhoff c, Maher Tabba d, John K Erban e a Taussig Cancer Institute of Cleveland Clinic, Department of Hematology/Oncology, Cleveland Clinic in Weston, FL, USA, b Department of Pathology, Tufts Medical Center Cancer Center & Tufts University School of Medicine, Boston, MA, USA, c Department of Surgery, Tufts Medical Center Cancer Center & Tufts University School of Medicine, Boston, MA, USA, d Division of Critical Care, Pulmonary and Sleep Medicine, Tufts Medical Center Cancer Center & Tufts University School of Medicine, Boston, MA, USA, e Division of Hematology/Oncology, Tufts Medical Center Cancer Center & Tufts University School of Medicine, Boston, MA, USA * Corresponding author at: Cleveland Clinic Florida, 2950 Cleveland Clinic Blvd., Weston, FL 33331, USA. Tel.: +1 954 659 5840; fax: +1 954 659 5810. Æ [email protected] Æ Received for publication 29 January 2014 Æ Accepted for publication 1 September 2014 Hematol Oncol Stem Cell Ther 2014; 7(4): 127–135 ª 2014 King Faisal Specialist Hospital & Research Centre. Published by Elsevier Ltd. All rights reserved. DOI: http://dx.doi.org/10.1016/j.hemonc.2014.09.003 Abstract Pulmonary alveolar proteinosis (PAP), characterized by deposition of intra-alveolar PAS positive protein and lipid rich material, is a rare cause of progressive respiratory failure first described by Rosen et al. in 1958. The intra-alveolar lipoproteinaceous material was subsequently proven to have been derived from pulmonary surfactant in 1980 by Singh et al. Levinson et al. also reported in 1958 the case of 19- year-old female with panmyelosis afflicted with a diffuse pulmonary disease characterized by filling of the alveoli with amorphous material described as ‘‘intra-alveolar coagulum’’. -

1700407.Full.Pdf
AGORA | CORRESPONDENCE Haemodynamically proven pulmonary hypertension in a patient with GATA2 deficiency-associated pulmonary alveolar proteinosis and fibrosis To the Editor: We read with great interest the article by BALLERIE et al. [1], which describes the occurrence of pulmonary alveolar proteinosis and fibrosis in a patient with GATA2 deficiency. Interestingly, our team recently managed a similar case, in which the interstitial lung disease eventually led to pulmonary hypertension (PH). A 41-year-old woman was referred to our tertiary care centre in 2014 for a suspected primary immunodeficiency. Her previous medical history included recurrent upper and lower respiratory tract infections, persisting cytopenias (including monocytopenia, and B-cell and natural killer cell lymphopenias), a disseminated Mycobacterium kansasii infection (in 2002) and a myelodysplastic syndrome (in 2008). She also displayed an interstitial lung disease that was first noted on a chest high-resolution computed tomography (HRCT) in 2002. A lung biopsy performed in 2007 showed histological lesions of pulmonary alveolar proteinosis and paraseptal fibrosis. Over the years, the pulmonary disease slowly progressed to involve the whole lung parenchyma. At the time of referral, chest HRCT displayed diffuse interlobular septal thickening, reticulations and ground-glass opacities of both lungs (figure 1a). The overall picture was compatible with the diagnosis of GATA2 deficiency, which was confirmed by molecular biology in October 2014. At the time of GATA2 deficiency diagnosis, the patient complained of a slowly progressive worsening of her dyspnoea and was staged in New York Heart Association functional class IIa. Physical examination showed no sign of right heart failure (including lower limb oedema). -

GATA2 Deficiency and Related Myeloid Neoplasms
Seminars in Hematology 54 (2017) 81–86 Contents lists available at ScienceDirect Seminars in Hematology journal homepage: www.elsevier.com/locate/enganabound GATA2 deficiency and related myeloid neoplasms Marcin W. Wlodarskia,b,n, Matthew Collinc, Marshall S. Horwitzd a Department of Pediatrics and Adolescent Medicine, Division of Pediatric Hematology and Oncology; Medical Center; Faculty of Medicine, University of Freiburg, Germany b German Cancer Consortium (DKTK), Freiburg, Germany and German Cancer Research Center (DKFZ), Heidelberg, Germany c Institute of Cellular Medicine, Newcastle University, Newcastle upon Tyne, United Kingdom; Northern Centre for Bone Marrow Transplantation, Newcastle upon Tyne Hospitals NHS Foundation Trust, Newcastle upon Tyne, United Kingdom d Department of Pathology, University of Washington School of Medicine, Seattle, WA, USA article info abstract Available online 19 May 2017 The GATA2 gene codes for a hematopoietic transcription factor that through its two zinc fingers (ZF) can occupy GATA-DNA motifs in a countless number of genes. It is crucial for the proliferation and Keywords: maintenance of hematopoietic stem cells. During the past 5 years, germline heterozygous mutations Genetic predisposition in GATA2 were reported in several hundred patients with various phenotypes ranging from mild GATA2 deficiency þ cytopenia to severe immunodeficiency involving B cells, natural killer cells, CD4 cells, monocytes and Familial MDS dendritic cells (MonoMAC/DCML), and myeloid neoplasia. Some patients additionally show -

Highly Variable Clinical Manifestations in a Large Family with a Novel GATA2 Mutation
Letters to the Editor 2247 interleukin-2 receptor expression and growth of leukemic Sezary cells. Leukemia 8 Chiarle R, Simmons WJ, Cai H, Dhall G, Zamo A, Raz R et al. Stat3 is required for 2001; 15: 787–793. ALK-mediated lymphomagenesis and provides a possible therapeutic target. 7 Ohgami RS, Ohgami JK, Pereira IT, Gitana G, Zehnder JL, Arber DA. Refining the Nat Med 2005; 11: 623–629. diagnosis of T-cell large granular lymphocytic leukemia by combining distinct patterns 9 Jing N, Tweardy DJ. Targeting Stat3 in cancer therapy. Anticancer Drugs 2005; 16: of antigen expression with T-cell clonality studies. Leukemia 2011; 25: 1439–1443. 601–607. Supplementary Information accompanies this paper on the Leukemia website (http://www.nature.com/leu) Highly variable clinical manifestations in a large family with a novel GATA2 mutation Leukemia (2013) 27, 2247–2248; doi:10.1038/leu.2013.105 non-Hodgkin lymphoma of the nasopharynx with rapidly progressive pancytopenia refractory to high-dose methylprednisolone and rituximab. The lymphoma was immunophenotypically CD3 and CD45 positive but CD4, CD8, CD20, CD30 and CD79a negative Recently, germline mutations in GATA2 have been described in and was classified as peripheral T-cell NHL, not otherwise specified. familial myelodysplastic syndrome/acute myeloid leukemia (MDS/ Bone marrow histology did not show marrow involvement of the AML) and in apparently related clinical conditions, designated lymphoma at the time of pancytopenia. DCML (dendritic cell-, mono- and lymphocytopenia), MonoMac The paternal grandmother of the index patient died at the age (monocytopenia and Mycobacterium avium complex infections) of 74 from AML. -

Dexi Disruption Depletes Gut Microbial Metabolites and Accelerates
bioRxiv preprint doi: https://doi.org/10.1101/393421; this version posted August 16, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Title: Dexi disruption depletes gut microbial metabolites and accelerates autoimmune diabetes LJ Davison, MD Wallace, C Preece, K Hughes, JA Todd, B Davies, CA O’Callaghan bioRxiv preprint doi: https://doi.org/10.1101/393421; this version posted August 16, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract: Non-coding genetic variants in the CLEC16A gene on human chromosome 16p13.13 are associated with risk of autoimmune diseases, including type 1 diabetes and multiple sclerosis. In this region, we previously identified DEXI, a candidate causal gene of unknown function, which alters the risk of type 1 diabetes, where the T1D predisposing allele is associated with lower DEXI expression. Here, we demonstrate by CRISPR mutagenesis in vivo and deep phenotyping that disrupted Dexi expression accelerates diabetes in the non-obese diabetic (NOD) mouse, a spontaneous model of autoimmune pancreatic beta-cell destruction. Mutant mice have increased serum IgM and IgA concentrations compared to wild-type NOD mice, as well as changes in both the gut microbiome and molecular metabolites associated with microbial metabolism. These findings suggest that the mechanism by which DEXI alters diabetes risk involves the composition and function of the microbiome and its impact on host metabolites. Such metabolites, including short chain fatty acids such as butyrate, have been shown to alter the activity of the immune cells involved in beta-cell destruction and susceptibility of the beta cells to autoimmune attack. -

Effects on Myeloid Cell Activation and Tumor
Int.J.Cancer:98,857–863(2002) PublicationoftheInternationalUnionAgainstCancer ©2002Wiley-Liss,Inc. DOI10.1002/ijc.10245 TUMOR-TARGETEDIMMUNECOMPLEXFORMATION:EFFECTSONMYELOID CELLACTIVATIONANDTUMOR-DIRECTEDIMMUNECELLMIGRATION 1 1 2,3 1 1 Bart-JanKROESEN ,*PamelaM.J.MCLAUGHLIN ,PetraH.L.SCHUILENGA-HUT ,SusanC.JACOBS ,GrietjeMOLEMA , 1 1 WijnandHELFRICH andLouF.M.H.DELEIJ 1DepartmentofPathologyandLaboratoryMedicine,UniversityHospitalGroningen,Groningen,TheNetherlands 2DepartmentofMedicalGenetics,UniversityofGroningen,Groningen,TheNetherlands 3GroningenUniversityInstituteforDrugExploration,Groningen,TheNetherlands Theeffectivenessofcellularimmunotherapyofsolidtu- inflammatoryreactionisbeinginitiated.TypeIIandtypeIII morsisoftenhamperedbythelackofspecificinfiltrationof inflammatoryreactionsareinducedbycytotoxicandcomplexed immuneeffectorcellsintothetumormass.Therefore,we antibodies,respectively,which,afterspecificornonspecificdep- studiedthepotentialoftumorantigen-specificantibodiesto ositionatcertainsites,activatemyeloidcellspresentinsituvia elicittumor-specificmyeloidcellactivation,toinduceor enhancetumorinfiltrationbyimmunecells.Tothisend,we Fc–FcRinteraction.Myeloidcellactivationcanbeinitiatedsolely developedaninvitromodelsystemusingthehumanmyeloid byFc–FcRinteraction,butothersignals,suchasthoseprovidedby celllineMonoMac-6.IncubationofIFN-␥–primedMono- componentsoflocallyactivatedcomplementand/orbycytokines Mac-6cellswithserum-opsonizedzymosanorEGP-2–di- suchasTNF-␣andIFN-␥,cangreatlyaugmentmyeloidcell rected,mouseIgG2a-opsonized,EGP-2-positivetumorcells -

Alveolar Proteinosis of Genetic Origins
SERIES RARE GENETIC ILDS Alveolar proteinosis of genetic origins Alice Hadchouel1,2,3, David Drummond1, Rola Abou Taam1, Muriel Lebourgeois1, Christophe Delacourt1,2,3 and Jacques de Blic1 Number 2 in the Series “Rare genetic interstitial lung diseases” Edited by Bruno Crestani and Raphaël Borie Affiliations: 1AP-HP, Hôpital Necker-Enfants Malades, Service de Pneumologie Pédiatrique, Centre de Référence pour les Maladies Respiratoires Rares de l’Enfant, Paris, France. 2INSERM U1151, Institut Necker Enfants Malades, Paris, France. 3Université de Paris, Faculté de Médecine, Paris, France. Correspondence: Alice Hadchouel, Service de Pneumologie et d’Allergologie Pédiatriques, Hôpital Universitaire Necker-Enfants Malades, 149 rue de Sèvres, 75743 Paris, France. E-mail: alice.hadchouel- [email protected] @ERSpublications Genetic PAP occurs in young children and is often associated with a poor prognosis. Next-generation sequencing panels represent an efficient diagnostic tool. Promising treatments are currently being developed for some specific entities. https://bit.ly/2TRsJd1 Cite this article as: Hadchouel A, Drummond D, Abou Taam R, et al. Alveolar proteinosis of genetic origins. Eur Respir Rev 2020; 29: 190187 [https://doi.org/10.1183/16000617.0187-2019]. ABSTRACT Pulmonary alveolar proteinosis (PAP) is a rare form of chronic interstitial lung disease, characterised by the intra-alveolar accumulation of lipoproteinaceous material. Numerous conditions can lead to its development. Whereas the autoimmune type is the main cause in adults, genetic defects account for a large part of cases in infants and children. Even if associated extra-respiratory signs may guide the clinician during diagnostic work-up, next-generation sequencing panels represent an efficient diagnostic tool. -

The Versatile Role of Alveolar Macrophages in Health and Disease
International Journal of Molecular Sciences Review Beyond “Big Eaters”: The Versatile Role of Alveolar Macrophages in Health and Disease Miriam Hetzel 1,2, Mania Ackermann 1,2,3 and Nico Lachmann 2,3,4,5,* 1 Institute of Experimental Hematology, Hannover Medical School, 30625 Hannover, Germany; [email protected] (M.H.); [email protected] (M.A.) 2 REBIRTH Research Center for Translational and Regenerative Medicine, Hannover Medical School, 30625 Hannover, Germany 3 Department of Pediatric Pneumology, Allergology and Neonatology, Hannover Medical School, 30625 Hannover, Germany 4 Biomedical Research in Endstage and Obstructive Lung Disease Hannover (BREATH), Member of the German Center for Lung Research (DZL), 30625 Hannover, Germany 5 Cluster of Excellence RESIST (EXC 2155), Hannover Medical School, 30625 Hannover, Germany * Correspondence: [email protected] Abstract: Macrophages act as immune scavengers and are important cell types in the homeostasis of various tissues. Given the multiple roles of macrophages, these cells can also be found as tissue resident macrophages tightly integrated into a variety of tissues in which they fulfill crucial and organ-specific functions. The lung harbors at least two macrophage populations: interstitial and alveolar macrophages, which occupy different niches and functions. In this review, we provide the latest insights into the multiple roles of alveolar macrophages while unraveling the distinct factors which can influence the ontogeny and function of these cells. Furthermore, we will high- light pulmonary diseases, which are associated with dysfunctional macrophages, concentrating on Citation: Hetzel, M.; Ackermann, M.; congenital diseases as well as pulmonary infections and impairment of immunological pathways. -

GATA2 Mutant Disease Phenotypes
OPEN Leukemia (2018) 32, 194–202 www.nature.com/leu ORIGINAL ARTICLE Differential effects on gene transcription and hematopoietic differentiation correlate with GATA2 mutant disease phenotypes C-E Chong1,2, P Venugopal1,2,3, PH Stokes4, YK Lee1,2, PJ Brautigan1,2, DTO Yeung1,2,5,6, M Babic1,2, GA Engler5, SW Lane7, M Klingler-Hoffmann3, JM Matthews4, RJ D'Andrea2,5,8, AL Brown1,2, CN Hahn1,2,6 and HS Scott1,2,3,6,8,9 Heterozygous GATA2 mutations underlie an array of complex hematopoietic and lymphatic diseases. Analysis of the literature reporting three recurrent GATA2 germline (g) mutations (gT354M, gR396Q and gR398W) revealed different phenotype tendencies. Although all three mutants differentially predispose to myeloid malignancies, there was no difference in leukemia-free survival for GATA2 patients. Despite intense interest, the molecular pathogenesis of GATA2 mutation is poorly understood. We functionally characterized a GATA2 mutant allelic series representing major disease phenotypes caused by germline and somatic (s) mutations in zinc finger 2 (ZF2). All GATA2 mutants, except for sL359V, displayed reduced DNA-binding affinity and transactivation compared with wild type (WT), which could be attributed to mutations of arginines critical for DNA binding or amino acids required for ZF2 domain structural integrity. Two GATA2 mutants (gT354M and gC373R) bound the key hematopoietic differentiation factor PU.1 more strongly than WT potentially perturbing differentiation via sequestration of PU.1. Unlike WT, all mutants failed to suppress colony formation and some mutants skewed cell fate to granulocytes, consistent with the monocytopenia phenotype seen in GATA2-related immunodeficiency disorders. These findings implicate perturbations of GATA2 function shaping the course of development of myeloid malignancy subtypes and strengthen complete or nearly complete haploinsufficiency for predisposition to lymphedema.