Single-Nucleotide Human Disease Mutation Inactivates a Blood- Regenerative GATA2 Enhancer
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Supplemental Material 1 (PDF)
Supplementary Figure 1. Murine and human GATA2 promoter alignment shows highly conserved regions. Brackets delimit the highly conserved regions in both genomes, as graphics obtained from ECR Browser show: large areas marked in red indicate highly similar intergenic sequences, repetitive sequences in green, exonic sequences in yellow, and intronic sequences in salmon color. Horizontal axis represents base positions in the genome and the vertical axis represents the percentage identity between the human and mouse genomes. GATA2 binding sites are indicated in red within the brackets, and they were located using the TFSearch, Match and MatInspector transcription factor binding site finder algorithms. Comparison of GATA2 binding sites between both genomes and several surrounding nucleotides is shown in the middle, where GATA2 binding site core sequences are shown in italic and underlined. Two GATA2 binding sites at -2.8 belonged to a highly conserved region spanning 389 bp, with 87.7% homology with the human GATA2 promoter. Similarly, other two GATA2 binding sites at -1.8 were located within a 282 bp region with 75.5% homology. Additionally, a third 142 bp region was taken into account containing the -2992 GATA2 binding site, which showed 68.3% homology. Supplementary Table I: List of reported germline GATA2 mutations and associated syndromes. Mutation (DNA)* Affected exon Mutation (protein) Mutation type Affected ZF Reference Cases/families Associated syndrome c.1-200_871+527del2033 4, 5 p.Met1_Ser290del In-frame deletion None 4,7,20,S1 2/1 -

ERG Dependence Distinguishes Developmental Control of Hematopoietic Stem Cell Maintenance from Hematopoietic Specification
Downloaded from genesdev.cshlp.org on September 28, 2021 - Published by Cold Spring Harbor Laboratory Press ERG dependence distinguishes developmental control of hematopoietic stem cell maintenance from hematopoietic specification Samir Taoudi,1,2,6 Thomas Bee,3 Adrienne Hilton,1 Kathy Knezevic,3 Julie Scott,4 Tracy A. Willson,1,2 Caitlin Collin,1 Tim Thomas,1,2 Anne K. Voss,1,2 Benjamin T. Kile,1,2 Warren S. Alexander,2,5 John E. Pimanda,3 and Douglas J. Hilton1,2 1Molecular Medicine Division, The Walter and Eliza Institute of Medical Research, Melbourne, Parkville, Victoria 3052, Australia; 2Department of Medical Biology, The University of Melbourne, Melbourne, Parkville, Victoria 3010, Australia; 3Lowy Cancer Research Centre, The Prince of Wales Clinical School, University of New South Wales, Sydney 2052, Australia; 4Microinjection Services, The Walter and Eliza Institute of Medical Research, Melbourne, Parkville, Victoria 3052, Australia; 5Cancer and Haematology Division, The Walter and Eliza Institute of Medical Research, Melbourne, Parkville, Victoria 3052, Australia Although many genes are known to be critical for early hematopoiesis in the embryo, it remains unclear whether distinct regulatory pathways exist to control hematopoietic specification versus hematopoietic stem cell (HSC) emergence and function. Due to their interaction with key regulators of hematopoietic commitment, particular interest has focused on the role of the ETS family of transcription factors; of these, ERG is predicted to play an important role in the initiation of hematopoiesis, yet we do not know if or when ERG is required. Using in vitro and in vivo models of hematopoiesis and HSC development, we provide strong evidence that ERG is at the center of a distinct regulatory program that is not required for hematopoietic specification or differentiation but is critical for HSC maintenance during embryonic development. -

Ubiquitin-Mediated Control of ETS Transcription Factors: Roles in Cancer and Development
International Journal of Molecular Sciences Review Ubiquitin-Mediated Control of ETS Transcription Factors: Roles in Cancer and Development Charles Ducker * and Peter E. Shaw * Queen’s Medical Centre, School of Life Sciences, University of Nottingham, Nottingham NG7 2UH, UK * Correspondence: [email protected] (C.D.); [email protected] (P.E.S.) Abstract: Genome expansion, whole genome and gene duplication events during metazoan evolution produced an extensive family of ETS genes whose members express transcription factors with a conserved winged helix-turn-helix DNA-binding domain. Unravelling their biological roles has proved challenging with functional redundancy manifest in overlapping expression patterns, a common consensus DNA-binding motif and responsiveness to mitogen-activated protein kinase signalling. Key determinants of the cellular repertoire of ETS proteins are their stability and turnover, controlled largely by the actions of selective E3 ubiquitin ligases and deubiquitinases. Here we discuss the known relationships between ETS proteins and enzymes that determine their ubiquitin status, their integration with other developmental signal transduction pathways and how suppression of ETS protein ubiquitination contributes to the malignant cell phenotype in multiple cancers. Keywords: E3 ligase complex; deubiquitinase; gene fusions; mitogens; phosphorylation; DNA damage 1. Introduction Citation: Ducker, C.; Shaw, P.E. Cell growth, proliferation and differentiation are complex, concerted processes that Ubiquitin-Mediated Control of ETS Transcription Factors: Roles in Cancer rely on careful regulation of gene expression. Control over gene expression is maintained and Development. Int. J. Mol. Sci. through signalling pathways that respond to external cellular stimuli, such as growth 2021, 22, 5119. https://doi.org/ factors, cytokines and chemokines, that invoke expression profiles commensurate with 10.3390/ijms22105119 diverse cellular outcomes. -
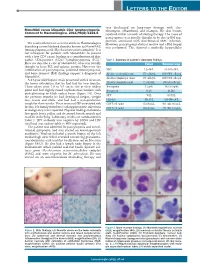
Letters to the Editor
LETTERS TO THE EDITOR was discharged on long-term therapy with clar - + MonoMAC versus idiopathic CD4 lymphocytopenia . ithromycin, ethambutol, and rifampin. His skin lesions Comment to Haematologica. 2011;96(8):1221-5 resolved within a month of starting therapy. The cause of pancytopenia was initially thought to be due to BM sup - pression associated with disseminated MAC infection. We read with interest a recent article in Haematologica However, pancytopenia did not resolve and a BM biopsy describing a novel defined disorder known as MonoMAC was performed. This showed a markedly hypocellular (monocytopenia with Mycobacterium avium complex). 1 It is not infrequent for patients with MonoMAC to present with a low CD4 count, leading to a consideration of idio - pathic CD4-positive (CD4 +) lymphocytopenia (ICL). 2 Table 1. Summary of patient’s laboratory findings. Here we describe a case of MonoMAC who was initially Patient Reference range thought to have ICL and aplastic anemia. However, the combination of pancytopenia, profound monocytopenia, WBC 1.2 ¥10 9/L 4.1-10.9 ¥10 9/L and bone marrow (BM) findings support a diagnosis of Absolute neutrophil count 876 cells/ mL 1400-6500 cells/ mL MonoMAC. Absolute lymphocyte count 144 cells/ mL 1200-3400 cells/ mL A 24-year old Hispanic male presented with 2 ulcers on the lower extremities that he had had for two months. Absolute monocyte count 12 cells/ mL 300-820 cells/ mL These ulcers were 1.0 to 3.5 cm in size at their widest Hemoglobin 5.2 g/dL 14.0-18.0 g/dL point and had slightly raised erythematous borders with Hematocrit 16.2% 40-54% pink-glistening to black eschar bases (Figure 1A). -

Inducible Transgene Expression in PDX Models
Liu et al. Biomarker Research (2020) 8:46 https://doi.org/10.1186/s40364-020-00226-z RESEARCH Open Access Inducible transgene expression in PDX models in vivo identifies KLF4 as a therapeutic target for B-ALL Wen-Hsin Liu1, Paulina Mrozek-Gorska2, Anna-Katharina Wirth1, Tobias Herold1,3, Larissa Schwarzkopf1, Dagmar Pich2, Kerstin Völse1, M. Camila Melo-Narváez2, Michela Carlet1, Wolfgang Hammerschmidt2,4 and Irmela Jeremias1,5,6* Abstract Background: Clinically relevant methods are not available that prioritize and validate potential therapeutic targets for individual tumors, from the vast amount of tumor descriptive expression data. Methods: We established inducible transgene expression in clinically relevant patient-derived xenograft (PDX) models in vivo to fill this gap. Results: With this technique at hand, we analyzed the role of the transcription factor Krüppel-like factor 4 (KLF4) in B-cell acute lymphoblastic leukemia (B-ALL) PDX models at different disease stages. In competitive preclinical in vivo trials, we found that re-expression of wild type KLF4 reduced the leukemia load in PDX models of B-ALL, with the strongest effects being observed after conventional chemotherapy in minimal residual disease (MRD). A nonfunctional KLF4 mutant had no effect on this model. The re-expression of KLF4 sensitized tumor cells in the PDX model towards systemic chemotherapy in vivo. It is of major translational relevance that azacitidine upregulated KLF4 levels in the PDX model and a KLF4 knockout reduced azacitidine-induced cell death, suggesting that azacitidine can regulate KLF4 re-expression. These results support the application of azacitidine in patients with B-ALL as a therapeutic option to regulate KLF4. -

Ten Commandments for a Good Scientist
Unravelling the mechanism of differential biological responses induced by food-borne xeno- and phyto-estrogenic compounds Ana María Sotoca Covaleda Wageningen 2010 Thesis committee Thesis supervisors Prof. dr. ir. Ivonne M.C.M. Rietjens Professor of Toxicology Wageningen University Prof. dr. Albertinka J. Murk Personal chair at the sub-department of Toxicology Wageningen University Thesis co-supervisor Dr. ir. Jacques J.M. Vervoort Associate professor at the Laboratory of Biochemistry Wageningen University Other members Prof. dr. Michael R. Muller, Wageningen University Prof. dr. ir. Huub F.J. Savelkoul, Wageningen University Prof. dr. Everardus J. van Zoelen, Radboud University Nijmegen Dr. ir. Toine F.H. Bovee, RIKILT, Wageningen This research was conducted under the auspices of the Graduate School VLAG Unravelling the mechanism of differential biological responses induced by food-borne xeno- and phyto-estrogenic compounds Ana María Sotoca Covaleda Thesis submitted in fulfillment of the requirements for the degree of doctor at Wageningen University by the authority of the Rector Magnificus Prof. dr. M.J. Kropff, in the presence of the Thesis Committee appointed by the Academic Board to be defended in public on Tuesday 14 September 2010 at 4 p.m. in the Aula Unravelling the mechanism of differential biological responses induced by food-borne xeno- and phyto-estrogenic compounds. Ana María Sotoca Covaleda Thesis Wageningen University, Wageningen, The Netherlands, 2010, With references, and with summary in Dutch. ISBN: 978-90-8585-707-5 “Caminante no hay camino, se hace camino al andar. Al andar se hace camino, y al volver la vista atrás se ve la senda que nunca se ha de volver a pisar” - Antonio Machado – A mi madre. -

Editorials & Perspectives
EDITORIALS & PERSPECTIVES Dendritic cell, monocyte, B and NK lymphoid deficiency defines the lost lineages of a new GATA-2 dependent myelodysplastic syndrome Venetia Bigley and Matthew Collin Institute of Cellular Medicine, Newcastle University, UK. E-mail: [email protected] doi:10.3324/haematol.2011.048355 (Related Original Article on page 1221) novel immunodeficiency syndrome has recently been International Prognostic Scoring System (IPSS). defined in young adults. Known variously as dendritic It is worth emphasizing at this point that DCML deficiency cell, monocyte, B and NK lymphoid (DCML deficien - patients may have significant risk to their health before addi - A1 2 2 cy), ‘autosomal dominant and sporadic monocytopenia’ or tional cytopenias evolve. Disseminated mycobacterial infec - ‘MonoMAC’ (monocytopenia with Mycobacterium avium com - tion, PAP and carcinoma in situ may all occur prior to the onset plex), 3 it is characterized by a composite mononuclear cell defi - of an obvious myelodysplastic syndrome, underscoring the ciency, atypical mycobacterial and viral infection, and progres - fact that immunodeficiency is a cardinal feature of the disor - sion to myelodysplasia and leukemia. Strikingly, a number of der. In the series of 4 subjects described in 2011, one had mild patients also develop pulmonary alveolar proteinosis (PAP). anemia (10.3 g/dL) and one mild thrombocytopenia Patients typically present in their 3 rd or 4 th decade, although his - (123 ¥10 9/L), yet 2 required hematopoietic stem cell transplan - torical blood counts may show pre-existing monocytopenia tation (one for refractory mycobacterial infection, one for PAP) for ten years or more. A substantial number of patients devel - and a third died of influenza H1N1 prior to transplantation. -

PBX3 and MEIS1 Cooperate in Hematopoietic Cells to Drive Acute
Published OnlineFirst January 8, 2016; DOI: 10.1158/0008-5472.CAN-15-1566 Cancer Molecular and Cellular Pathobiology Research PBX3 and MEIS1 Cooperate in Hematopoietic Cells to Drive Acute Myeloid Leukemias Characterized by a Core Transcriptome of the MLL-Rearranged Disease Zejuan Li1, Ping Chen1, Rui Su2, Chao Hu1,2,3, Yuanyuan Li1, Abdel G. Elkahloun4, Zhixiang Zuo1,2, Sandeep Gurbuxani5, Stephen Arnovitz1, Hengyou Weng1,2, Yungui Wang1,2,3, Shenglai Li1, Hao Huang1, Mary Beth Neilly1, Gang Greg Wang6, Xi Jiang1,2, Paul P. Liu4, Jie Jin3, and Jianjun Chen1,2 Abstract Overexpression of HOXA/MEIS1/PBX3 homeobox genes is sion profiling of hematopoietic cells demonstrated that PBX3/ the hallmark of mixed lineage leukemia (MLL)-rearranged MEIS1 overexpression, but not HOXA9/MEIS1, HOXA9/PBX3, acute myeloid leukemia (AML). HOXA9 and MEIS1 are con- or HOXA9 overexpression, recapitulated the MLL-fusion–medi- sideredtobethemostcriticaltargetsofMLLfusionsandtheir ated core transcriptome, particularly upregulation of the endog- coexpression rapidly induces AML. MEIS1 and PBX3 are not enous Hoxa genes. Disruption of the binding between MEIS1 individually able to transform cells and were therefore hypoth- andPBX3diminishedPBX3/MEIS1–mediated cell transforma- esized to function as cofactors of HOXA9. However, in this tion and HOX gene upregulation. Collectively, our studies study, we demonstrate that coexpression of PBX3 and MEIS1 strongly implicate the PBX3/MEIS1 interaction as a driver of (PBX3/MEIS1), without ectopic expression of a HOX gene, is cell transformation and leukemogenesis, and suggest that this sufficient for transformation of normal mouse hematopoietic axis may play a critical role in the regulation of the core stem/progenitor cells in vitro.Moreover,PBX3/MEIS1 overex- transcriptional programs activated in MLL-rearranged and pression also caused AML in vivo, with a leukemic latency HOX-overexpressing AML. -

Secondary Pulmonary Alveolar Proteinosis in Hematologic
review Secondary pulmonary alveolar proteinosis in hematologic malignancies Chakra P Chaulagain a,*, Monika Pilichowska b, Laurence Brinckerhoff c, Maher Tabba d, John K Erban e a Taussig Cancer Institute of Cleveland Clinic, Department of Hematology/Oncology, Cleveland Clinic in Weston, FL, USA, b Department of Pathology, Tufts Medical Center Cancer Center & Tufts University School of Medicine, Boston, MA, USA, c Department of Surgery, Tufts Medical Center Cancer Center & Tufts University School of Medicine, Boston, MA, USA, d Division of Critical Care, Pulmonary and Sleep Medicine, Tufts Medical Center Cancer Center & Tufts University School of Medicine, Boston, MA, USA, e Division of Hematology/Oncology, Tufts Medical Center Cancer Center & Tufts University School of Medicine, Boston, MA, USA * Corresponding author at: Cleveland Clinic Florida, 2950 Cleveland Clinic Blvd., Weston, FL 33331, USA. Tel.: +1 954 659 5840; fax: +1 954 659 5810. Æ [email protected] Æ Received for publication 29 January 2014 Æ Accepted for publication 1 September 2014 Hematol Oncol Stem Cell Ther 2014; 7(4): 127–135 ª 2014 King Faisal Specialist Hospital & Research Centre. Published by Elsevier Ltd. All rights reserved. DOI: http://dx.doi.org/10.1016/j.hemonc.2014.09.003 Abstract Pulmonary alveolar proteinosis (PAP), characterized by deposition of intra-alveolar PAS positive protein and lipid rich material, is a rare cause of progressive respiratory failure first described by Rosen et al. in 1958. The intra-alveolar lipoproteinaceous material was subsequently proven to have been derived from pulmonary surfactant in 1980 by Singh et al. Levinson et al. also reported in 1958 the case of 19- year-old female with panmyelosis afflicted with a diffuse pulmonary disease characterized by filling of the alveoli with amorphous material described as ‘‘intra-alveolar coagulum’’. -

RUNX1 Together with a Compendium of Hematopoietic Regulators, Chromatin Modifiers and Basal Transcr
Leukemia (2014) 28, 770–778 OPEN & 2014 Macmillan Publishers Limited All rights reserved 0887-6924/14 www.nature.com/leu ORIGINAL ARTICLE CBFB–MYH11/RUNX1 together with a compendium of hematopoietic regulators, chromatin modifiers and basal transcription factors occupies self-renewal genes in inv(16) acute myeloid leukemia A Mandoli1, AA Singh1, PWTC Jansen2, ATJ Wierenga3,4, H Riahi1, G Franci5, K Prange1, S Saeed1, E Vellenga3, M Vermeulen2, HG Stunnenberg1 and JHA Martens1 Different mechanisms for CBFb–MYH11 function in acute myeloid leukemia with inv(16) have been proposed such as tethering of RUNX1 outside the nucleus, interference with transcription factor complex assembly and recruitment of histone deacetylases, all resulting in transcriptional repression of RUNX1 target genes. Here, through genome-wide CBFb–MYH11-binding site analysis and quantitative interaction proteomics, we found that CBFb–MYH11 localizes to RUNX1 occupied promoters, where it interacts with TAL1, FLI1 and TBP-associated factors (TAFs) in the context of the hematopoietic transcription factors ERG, GATA2 and PU.1/SPI1 and the coregulators EP300 and HDAC1. Transcriptional analysis revealed that upon fusion protein knockdown, a small subset of the CBFb–MYH11 target genes show increased expression, confirming a role in transcriptional repression. However, the majority of CBFb–MYH11 target genes, including genes implicated in hematopoietic stem cell self-renewal such as ID1, LMO1 and JAG1, are actively transcribed and repressed upon fusion protein knockdown. Together these results suggest an essential role for CBFb–MYH11 in regulating the expression of genes involved in maintaining a stem cell phenotype. Leukemia (2014) 28, 770–778; doi:10.1038/leu.2013.257 Keywords: CBFb–MYH11; RUNX1; histone acetylation; acute myeloid leukemia; inv(16) INTRODUCTION Heterozygous Cbfb-Myh11 knock-in mice are embryonic lethal, Core-binding transcription factors (CBFs) have roles in stem cell with definitive hematopoiesis blocked at the stem-cell level. -

The Histone Methyltransferase MMSET/WHSC1 Activates TWIST1 to Promote an Epithelial–Mesenchymal Transition and Invasive Properties of Prostate Cancer
Oncogene (2013) 32, 2882–2890 & 2013 Macmillan Publishers Limited All rights reserved 0950-9232/13 www.nature.com/onc ORIGINAL ARTICLE The histone methyltransferase MMSET/WHSC1 activates TWIST1 to promote an epithelial–mesenchymal transition and invasive properties of prostate cancer T Ezponda1, R Popovic1, MY Shah1, E Martinez-Garcia1, Y Zheng2, D-J Min1, C Will1, A Neri3, NL Kelleher1,2,JYu1 and JD Licht1 Epigenetic deregulation of gene expression has a role in the initiation and progression of prostate cancer (PCa). The histone methyltransferase MMSET/WHSC1 (Multiple Myeloma SET domain) is overexpressed in a number of metastatic tumors, but its mechanism of action has not been defined. In this work, we found that PCa cell lines expressed significantly higher levels of MMSET compared with immortalized, non-transformed prostate cells. Knockdown experiments showed that, in metastatic PCa cell lines, dimethylation of lysine 36 and trimethylation of lysine 27 on histone H3 (H3K36me2 and H3K27me3, respectively) depended on MMSET expression, whereas depletion of MMSET in benign prostatic cells did not affect chromatin modifications. Knockdown of MMSET in DU145 and PC-3 tumor cells decreased cell proliferation, colony formation in soft agar and strikingly diminished cell migration and invasion. Conversely, overexpression of MMSET in immortalized, non-transformed RWPE-1 cells promoted cell migration and invasion, accompanied by an epithelial–mesenchymal transition (EMT). Among a panel of EMT-promoting genes analyzed, TWIST1 expression was strongly activated in response to MMSET. Chromatin immunoprecipitation analysis demonstrated that MMSET binds to the TWIST1 locus and leads to an increase in H3K36me2, suggesting a direct role of MMSET in the regulation of this gene. -

GATA3-Controlled Nucleosome Eviction Drives MYC Enhancer Activity in T-Cell Development and Leukemia
Published OnlineFirst September 13, 2019; DOI: 10.1158/2159-8290.CD-19-0471 RESEARCH ARTICLE GATA3-Controlled Nucleosome Eviction Drives MYC Enhancer Activity in T-cell Development and Leukemia Laura Belver1, Alexander Y. Yang1, Robert Albero1, Daniel Herranz2,3, Francesco G. Brundu4, S. Aidan Quinn1, Pablo Pérez-Durán1, Silvia Álvarez1, Francesca Gianni1, Marissa Rashkovan1, Devya Gurung1, Pedro P. Rocha5, Ramya Raviram6,7, Clara Reglero1, Jose R. Cortés1, Anisha J. Cooke1, Agnieszka A. Wendorff1, Valentina Cordó8, Jules P. Meijerink8, Raúl Rabadan4,9, and Adolfo A. Ferrando1,4,10,11 ABSTRACT Long-range enhancers govern the temporal and spatial control of gene express ion; however, the mechanisms that regulate enhancer activity during normal and malig- nant development remain poorly understood. Here, we demonstrate a role for aberrant chromatin acces- sibility in the regulation of MYC expression in T-cell lymphoblastic leukemia (T-ALL). Central to this process, the NOTCH1-MYC enhancer (N-Me), a long-range T cell–specificMYC enhancer, shows dynamic changes in chromatin accessibility during T-cell specification and maturation and an aberrant high degree of chromatin accessibility in mouse and human T-ALL cells. Mechanistically, we demonstrate that GATA3- driven nucleosome eviction dynamically modulates N-Me enhancer activity and is strictly required for NOTCH1-induced T-ALL initiation and maintenance. These results directly implicate aberrant regulation of chromatin accessibility at oncogenic enhancers as a mechanism of leukemic transformation. SIGNIFICANCE: MYC is a major effector of NOTCH1 oncogenic programs in T-ALL. Here, we show a major role for GATA3-mediated enhancer nucleosome eviction as a driver of MYC expression and leuke- mic transformation.