Differential Gene Expression in Human Abdominal Aortic Aneurysm and Aortic Occlusive Disease
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

WO 2016/147053 Al 22 September 2016 (22.09.2016) P O P C T
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date WO 2016/147053 Al 22 September 2016 (22.09.2016) P O P C T (51) International Patent Classification: (71) Applicant: RESVERLOGIX CORP. [CA/CA]; 300, A61K 31/551 (2006.01) A61P 37/02 (2006.01) 4820 Richard Road Sw, Calgary, AB, T3E 6L1 (CA). A61K 31/517 (2006.01) C07D 239/91 (2006.01) (72) Inventors: WASIAK, Sylwia; 431 Whispering Water (21) International Application Number: Trail, Calgary, AB, T3Z 3V1 (CA). KULIKOWSKI, PCT/IB20 16/000443 Ewelina, B.; 31100 Swift Creek Terrace, Calgary, AB, T3Z 0B7 (CA). HALLIDAY, Christopher, R.A.; 403 (22) International Filing Date: 138-18th Avenue SE, Calgary, AB, T2G 5P9 (CA). GIL- 10 March 2016 (10.03.2016) HAM, Dean; 249 Scenic View Close NW, Calgary, AB, (25) Filing Language: English T3L 1Y5 (CA). (26) Publication Language: English (81) Designated States (unless otherwise indicated, for every kind of national protection available): AE, AG, AL, AM, (30) Priority Data: AO, AT, AU, AZ, BA, BB, BG, BH, BN, BR, BW, BY, 62/132,572 13 March 2015 (13.03.2015) US BZ, CA, CH, CL, CN, CO, CR, CU, CZ, DE, DK, DM, 62/264,768 8 December 2015 (08. 12.2015) US DO, DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, [Continued on nextpage] (54) Title: COMPOSITIONS AND THERAPEUTIC METHODS FOR THE TREATMENT OF COMPLEMENT-ASSOCIATED DISEASES (57) Abstract: The invention comprises methods of modulating the complement cascade in a mammal and for treating and/or preventing diseases and disorders as sociated with the complement pathway by administering a compound of Formula I or Formula II, such as, for example, 2-(4-(2-hydroxyethoxy)-3,5-dimethylphenyl)- 5,7-dimethoxyquinazolin-4(3H)-one or a pharmaceutically acceptable salt thereof. -

Mutation in the COX4I1 Gene Is Associated with Short Stature, Poor Weight Gain and Increased Chromosomal Breaks, Simulating Fanconi Anemia
European Journal of Human Genetics (2017) 25, 1142–1146 & 2017 Macmillan Publishers Limited, part of Springer Nature. All rights reserved 1018-4813/17 www.nature.com/ejhg ARTICLE Mutation in the COX4I1 gene is associated with short stature, poor weight gain and increased chromosomal breaks, simulating Fanconi anemia Bassam Abu-Libdeh*,1,4, Liza Douiev2,3,4, Sarah Amro1, Maher Shahrour1, Asaf Ta-Shma2, Chaya Miller2,3, Orly Elpeleg2 and Ann Saada*,2,3 We describe a novel autosomal recessive form of mitochondrial disease in a child with short stature, poor weight gain, and mild dysmorphic features with highly suspected Fanconi anemia due to a mutation in COX4I1 gene. Whole Exome Sequencing was performed then followed by Sanger confirmation, identified a K101N mutation in COX4I1, segregating with the disease. This nuclear gene encodes the common isoform of cytochrome c oxidase (COX) subunit 4 (COX 4-1), an integral regulatory part of COX (respiratory chain complex IV) the terminal electron acceptor of the mitochondrial respiratory chain. The patient’s fibroblasts disclosed decreased COX activity, impaired ATP production, elevated ROS production, decreased expression of COX4I1 mRNA and undetectable (COX4) protein. COX activity and ATP production were restored by lentiviral transfection with the wild-type gene. Our results demonstrate the first human mutation in the COX4I1 gene linked to diseases and confirm its role in the pathogenesis. Thus COX4I1 mutations should be considered in any patient with features suggestive of this diagnosis. European Journal of Human Genetics (2017) 25, 1142–1146; doi:10.1038/ejhg.2017.112; published online 2 August 2017 INTRODUCTION normal vaginal delivery with birth weight of 2.8 kg after an uneventful Mitochondrial cytochrome c oxidase (COX, complex IV) is the terminal pregnancy. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

Structure of the Intact 14-Subunit Human Cytochrome C Oxidase
www.nature.com/cr www.cell-research.com ARTICLE Structure of the intact 14-subunit human cytochrome c oxidase Shuai Zong 1, Meng Wu1, Jinke Gu1, Tianya Liu1, Runyu Guo1 and Maojun Yang1,2 Respiration is one of the most basic features of living organisms, and the electron transport chain complexes are probably the most complicated protein system in mitochondria. Complex-IV is the terminal enzyme of the electron transport chain, existing either as randomly scattered complexes or as a component of supercomplexes. NDUFA4 was previously assumed as a subunit of complex-I, but recent biochemical data suggested it may be a subunit of complex-IV. However, no structural evidence supporting this notion was available till now. Here we obtained the 3.3 Å resolution structure of complex-IV derived from the human supercomplex I1III2IV1 and assigned the NDUFA4 subunit into complex-IV. Intriguingly, NDUFA4 lies exactly at the dimeric interface observed in previously reported crystal structures of complex-IV homodimer which would preclude complex- IV dimerization. Combining previous structural and biochemical data shown by us and other groups, we propose that the intact complex-IV is a monomer containing 14 subunits. Cell Research (2018) 28:1026–1034; https://doi.org/10.1038/s41422-018-0071-1 INTRODUCTION states under physiological conditions, either being assembled into Mitochondria are critical to many cellular activities. Respiration is supercomplexes or freely scattered on mitochondrial inner the central function of mitochondria, and is exquisitely regulated membrane.36 NDUFA4 was originally considered as a subunit of in multiple ways in response to varying cell conditions.1–3 The Complex-I37 but was proposed to belong to Complex-IV recently.38 assembly of respiratory chain complexes, Complex I–IV (CI, NADH: However, the precise location of NDUFA4 in CIV remains unknown, ubiquinone oxidoreductase; CII, succinate:ubiquinone oxidoreduc- and thus this notion still lacks structural support. -
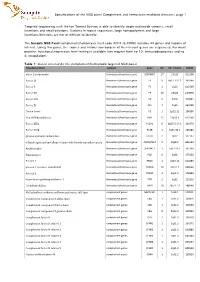
Specifications of the NGS Panel Complement and Hemostasis Mediated Diseases| Page 1
Specifications of the NGS panel Complement and hemostasis mediated diseases| page 1 Targeted sequencing with the Ion Torrent System is able to identify single nucleotide variants, small insertions and small deletions. Variants in repeat sequences, large homopolymers and large insertions/deletions are not or difficult to identify. The Sanquin NGS Panel complement/hemostasis (test code X001 to X006) includes 44 genes and regions of interest. Using this panel, the exones and intron/exon borders of the relevant genes are sequenced. For most proteins, functional/expression level testing is available (see request form no 10: immunodiagnostics and no 4: coagulation). Table 1: Genes covered by the complement/hemostasis targeted NGS panel. Encoded protein Context Gene Chr Chr. Positie OMIM alpha 2-antiplasmin Hemostasis/trombosis gene SERPINF2 17 17p13 613168 Factor IX Hemostasis/trombosis gene F9 X Xq27.1-27.2 300746 Factor V Hemostasis/trombosis gene F5 1 1q23 612309 Factor VII Hemostasis/trombosis gene F7 13 13q34 613878 Factor VIII Hemostasis/trombosis gene F8 X Xq28 300841 Factor XI Hemostasis/trombosis gene F11 4 4q35 264900 Tissue factor Hemostasis/trombosis gene F3 1 1p22-21 134390 Von Willebrand factor Hemostasis/trombosis gene VWF 12 12p13.3 613160 Factor XIIIa Hemostasis/trombosis gene F13A1 6 6p25.3-24.3 134570 Factor XIIIb Hemostasis/trombosis gene F13B 1 1q31-32.1 134580 gamma-glutamyl carboxylase Hemostasis/trombosis gene GGCX 2 2p12 137167 a Disintergrin and metalloproteinase wih thrombospondin repeats Hemostasis/trombosis gene -

A High-Resolution Genome-Wide CRISPR/Cas9 Viability Screen
RESEARCH ARTICLE crossm A High-Resolution Genome-Wide CRISPR/Cas9 Viability Screen Reveals Structural Features and Contextual Diversity of the Human Cell-Essential Proteome Downloaded from Thierry Bertomeu,a Jasmin Coulombe-Huntington,a Andrew Chatr-aryamontri,a Karine G. Bourdages,a Etienne Coyaud,b Brian Raught,b Yu Xia,c Mike Tyersa aInstitute for Research in Immunology and Cancer, Department of Medicine, University of Montreal, Montreal, Quebec, Canada bPrincess Margaret Cancer Centre, University Health Network, Toronto, Ontario, Canada cDepartment of Bioengineering, McGill University, Montreal, Quebec, Canada ABSTRACT To interrogate genes essential for cell growth, proliferation and survival http://mcb.asm.org/ in human cells, we carried out a genome-wide clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9 screen in a B-cell lymphoma line using a custom extended-knockout (EKO) library of 278,754 single-guide RNAs (sgRNAs) that tar- geted 19,084 RefSeq genes, 20,852 alternatively spliced exons, and 3,872 hypotheti- cal genes. A new statistical analysis tool called robust analytics and normalization for knockout screens (RANKS) identified 2,280 essential genes, 234 of which were unique. Individual essential genes were validated experimentally and linked to ribo- some biogenesis and stress responses. Essential genes exhibited a bimodal distribu- on December 15, 2018 by guest tion across 10 different cell lines, consistent with a continuous variation in essential- ity as a function of cell type. Genes essential in more lines had more severe fitness defects and encoded the evolutionarily conserved structural cores of protein com- plexes, whereas genes essential in fewer lines formed context-specific modules and encoded subunits at the periphery of essential complexes. -

Role of Cytochrome C Oxidase Nuclear-Encoded Subunits in Health and Disease
Physiol. Res. 69: 947-965, 2020 https://doi.org/10.33549/physiolres.934446 REVIEW Role of Cytochrome c Oxidase Nuclear-Encoded Subunits in Health and Disease Kristýna ČUNÁTOVÁ1, David PAJUELO REGUERA1, Josef HOUŠTĚK1, Tomáš MRÁČEK1, Petr PECINA1 1Department of Bioenergetics, Institute of Physiology, Czech Academy of Sciences, Prague, Czech Republic Received February 2, 2020 Accepted September 13, 2020 Epub Ahead of Print November 2, 2020 Summary [email protected] and Tomáš Mráček, Department of Cytochrome c oxidase (COX), the terminal enzyme of Bioenergetics, Institute of Physiology CAS, Vídeňská 1083, 142 mitochondrial electron transport chain, couples electron transport 20 Prague 4, Czech Republic. E-mail: [email protected] to oxygen with generation of proton gradient indispensable for the production of vast majority of ATP molecules in mammalian Cytochrome c oxidase cells. The review summarizes current knowledge of COX structure and function of nuclear-encoded COX subunits, which may Energy demands of mammalian cells are mainly modulate enzyme activity according to various conditions. covered by ATP synthesis carried out by oxidative Moreover, some nuclear-encoded subunits possess tissue-specific phosphorylation apparatus (OXPHOS) located in the and development-specific isoforms, possibly enabling fine-tuning central bioenergetic organelle, mitochondria. OXPHOS is of COX function in individual tissues. The importance of nuclear- composed of five multi-subunit complexes embedded in encoded subunits is emphasized by recently discovered the inner mitochondrial membrane (IMM). Electron pathogenic mutations in patients with severe mitopathies. In transport from reduced substrates of complexes I and II to addition, proteins substoichiometrically associated with COX were cytochrome c oxidase (COX, complex IV, CIV) is found to contribute to COX activity regulation and stabilization of achieved by increasing redox potential of individual the respiratory supercomplexes. -

Human Mitochondrial Pathologies of the Respiratory Chain and ATP Synthase: Contributions from Studies of Saccharomyces Cerevisiae
life Review Human Mitochondrial Pathologies of the Respiratory Chain and ATP Synthase: Contributions from Studies of Saccharomyces cerevisiae Leticia V. R. Franco 1,2,* , Luca Bremner 1 and Mario H. Barros 2 1 Department of Biological Sciences, Columbia University, New York, NY 10027, USA; [email protected] 2 Department of Microbiology,Institute of Biomedical Sciences, Universidade de Sao Paulo, Sao Paulo 05508-900, Brazil; [email protected] * Correspondence: [email protected] Received: 27 October 2020; Accepted: 19 November 2020; Published: 23 November 2020 Abstract: The ease with which the unicellular yeast Saccharomyces cerevisiae can be manipulated genetically and biochemically has established this organism as a good model for the study of human mitochondrial diseases. The combined use of biochemical and molecular genetic tools has been instrumental in elucidating the functions of numerous yeast nuclear gene products with human homologs that affect a large number of metabolic and biological processes, including those housed in mitochondria. These include structural and catalytic subunits of enzymes and protein factors that impinge on the biogenesis of the respiratory chain. This article will review what is currently known about the genetics and clinical phenotypes of mitochondrial diseases of the respiratory chain and ATP synthase, with special emphasis on the contribution of information gained from pet mutants with mutations in nuclear genes that impair mitochondrial respiration. Our intent is to provide the yeast mitochondrial specialist with basic knowledge of human mitochondrial pathologies and the human specialist with information on how genes that directly and indirectly affect respiration were identified and characterized in yeast. Keywords: mitochondrial diseases; respiratory chain; yeast; Saccharomyces cerevisiae; pet mutants 1. -

A Case of Meningococcal Sepsis and Meningitis with Complement 7
Case Report Infection & http://dx.doi.org/10.3947/ic.2013.45.1.94 Infect Chemother 2013;45(1):94-98 Chemotherapy pISSN 2093-2340 · eISSN 2092-6448 A Case of Meningococcal Sepsis and Meningitis with Complement 7 Deficiency in a Military Trainee Sung Hoon Sim1*, Jung Yeon Heo2*, Eui-Chong Kim3, and Kang-Won Choe2 1Department of Internal Medicine, Seoul National University Hospital, Seoul; 2Department of Internal Medicine, Armed Forces Capital Hospital, Seongnam; 3Department of Laboratory Medicine, Seoul National University Hospital, Seoul, Korea Complement component 7 (C7) deficiency leads to the loss of complement lytic function, and affected patients show in- creased susceptibility to encapsulated organisms infection, especially Neisseria meningitidis. Recently, we have experi- enced a 20-year-old military trainee with meningococcal sepsis and meningitis who was diagnosed as having C7 deficiency based upon the undetectable serum C7 protein on radioimmunoassay. This case emphasizes that although C7 deficiency is rare immune disorder, it is important to be aware of possibility about late complement deficiency among patients who pres- ent with meningococcal disease. Key Words: Neisseria meningitidis, Complement 7 deficiency, Meningitis, Sepsis Introduction of 20 and college freshmen residing in the dormitories are a well-known high risk group. The incidence of meningococcal Humans are the only host for Neisseria meningitidis which disease increases as those high risk age groups from different colonizes the nasopharynx and it is transmitted from human geological areas start communal living [2]. to human by infected respiratory secretions or saliva via air- Apart from these environmental factors, host factors such as borne respiratory droplets. -

Electron Transport Chain Activity Is a Predictor and Target for Venetoclax Sensitivity in Multiple Myeloma
ARTICLE https://doi.org/10.1038/s41467-020-15051-z OPEN Electron transport chain activity is a predictor and target for venetoclax sensitivity in multiple myeloma Richa Bajpai1,7, Aditi Sharma 1,7, Abhinav Achreja2,3, Claudia L. Edgar1, Changyong Wei1, Arusha A. Siddiqa1, Vikas A. Gupta1, Shannon M. Matulis1, Samuel K. McBrayer 4, Anjali Mittal3,5, Manali Rupji 6, Benjamin G. Barwick 1, Sagar Lonial1, Ajay K. Nooka 1, Lawrence H. Boise 1, Deepak Nagrath2,3,5 & ✉ Mala Shanmugam 1 1234567890():,; The BCL-2 antagonist venetoclax is highly effective in multiple myeloma (MM) patients exhibiting the 11;14 translocation, the mechanistic basis of which is unknown. In evaluating cellular energetics and metabolism of t(11;14) and non-t(11;14) MM, we determine that venetoclax-sensitive myeloma has reduced mitochondrial respiration. Consistent with this, low electron transport chain (ETC) Complex I and Complex II activities correlate with venetoclax sensitivity. Inhibition of Complex I, using IACS-010759, an orally bioavailable Complex I inhibitor in clinical trials, as well as succinate ubiquinone reductase (SQR) activity of Complex II, using thenoyltrifluoroacetone (TTFA) or introduction of SDHC R72C mutant, independently sensitize resistant MM to venetoclax. We demonstrate that ETC inhibition increases BCL-2 dependence and the ‘primed’ state via the ATF4-BIM/NOXA axis. Further, SQR activity correlates with venetoclax sensitivity in patient samples irrespective of t(11;14) status. Use of SQR activity in a functional-biomarker informed manner may better select for MM patients responsive to venetoclax therapy. 1 Department of Hematology and Medical Oncology, Winship Cancer Institute, School of Medicine, Emory University, Atlanta, GA, USA. -

Observation of Novel COX20 Mutations Related to Autosomal Recessive Axonal Neuropathy and Static Encephalopathy
Human Genetics (2019) 138:749–756 https://doi.org/10.1007/s00439-019-02026-4 ORIGINAL INVESTIGATION Observation of novel COX20 mutations related to autosomal recessive axonal neuropathy and static encephalopathy Hongliang Xu1 · Tuo Ji1 · Yajun Lian1 · Shuya Wang2 · Xin Chen1 · Shuang Li1 · Yuhui Yin3 · Xiubing Dong4 Received: 21 February 2019 / Accepted: 7 May 2019 / Published online: 11 May 2019 © Springer-Verlag GmbH Germany, part of Springer Nature 2019 Abstract Cytochrome c oxidase 20 (COX20)/FAM36A encodes a conserved protein that is important for the assembly of COX, com- plex IV of the mitochondrial respiratory chain. A homozygous mutation (p.Thr52Pro) in COX20 gene has been previously described to cause muscle hypotonia and ataxia. In this study, we describe two patients from a non-consanguineous family exhibiting autosomal recessive sensory-dominant axonal neuropathy and static encephalopathy. The whole-exome sequencing analysis revealed that both patients harbored compound heterozygous mutations (p.Lys14Arg and p.Trp74Cys) of COX20 gene. The pathogenicity of the variants was further supported by morphological alternations of mitochondria observed in sural nerve and decreased COX20 protein level of peripheral blood leucocytes derived from the patients. In conclusion, COX20 might be considered as a candidate gene for the complex inherited disease. This observation broadens the clinical and genetic spectrum of COX20-related disease. However, due to the limitation of a single-family study, additional cases and studies are defnitely needed to further confrm the association. Introduction a frequent cause of oxidative phosphorylation disorders in humans. Patients sufering from COX-related mitochondrial Cytochrome c oxidase 20 (COX20, also known as FAM36A) diseases present with heterogeneous clinical phenotypes encodes a conserved protein that is important for the assem- ranging from encephalomyopathy, hypertrophic cardio- bly of COX, complex IV of the mitochondrial respiratory myopathy and liver disease to Leigh’s syndrome (Bourens chain (Bourens et al. -

Transcriptomic and Proteomic Landscape of Mitochondrial
TOOLS AND RESOURCES Transcriptomic and proteomic landscape of mitochondrial dysfunction reveals secondary coenzyme Q deficiency in mammals Inge Ku¨ hl1,2†*, Maria Miranda1†, Ilian Atanassov3, Irina Kuznetsova4,5, Yvonne Hinze3, Arnaud Mourier6, Aleksandra Filipovska4,5, Nils-Go¨ ran Larsson1,7* 1Department of Mitochondrial Biology, Max Planck Institute for Biology of Ageing, Cologne, Germany; 2Department of Cell Biology, Institute of Integrative Biology of the Cell (I2BC) UMR9198, CEA, CNRS, Univ. Paris-Sud, Universite´ Paris-Saclay, Gif- sur-Yvette, France; 3Proteomics Core Facility, Max Planck Institute for Biology of Ageing, Cologne, Germany; 4Harry Perkins Institute of Medical Research, The University of Western Australia, Nedlands, Australia; 5School of Molecular Sciences, The University of Western Australia, Crawley, Australia; 6The Centre National de la Recherche Scientifique, Institut de Biochimie et Ge´ne´tique Cellulaires, Universite´ de Bordeaux, Bordeaux, France; 7Department of Medical Biochemistry and Biophysics, Karolinska Institutet, Stockholm, Sweden Abstract Dysfunction of the oxidative phosphorylation (OXPHOS) system is a major cause of human disease and the cellular consequences are highly complex. Here, we present comparative *For correspondence: analyses of mitochondrial proteomes, cellular transcriptomes and targeted metabolomics of five [email protected] knockout mouse strains deficient in essential factors required for mitochondrial DNA gene (IKu¨ ); expression, leading to OXPHOS dysfunction. Moreover,