UC Berkeley UC Berkeley Previously Published Works
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

History and Philosophy of Systematic Biology
History and Philosophy of Systematic Biology Bock, W. J. (1973) Philosophical foundations of classical evolutionary classification Systematic Zoology 22: 375-392 Part of a general symposium on "Contemporary Systematic Philosophies," there are some other interesting papers here. Brower, A. V. Z. (2000) Evolution Is Not a Necessary Assumption of Cladistics Cladistics 16: 143- 154 Dayrat, Benoit (2005) Ancestor-descendant relationships and the reconstruction of the Tree of Lif Paleobiology 31: 347-353 Donoghue, M.J. and J.W. Kadereit (1992) Walter Zimmermann and the growth of phylogenetic theory Systematic Biology 41: 74-84 Faith, D. P. and J. W. H. Trueman (2001) Towards an inclusive philosophy for phylogenetic inference Systematic Biology 50: 331-350 Gaffney, E. S. (1979) An introduction to the logic of phylogeny reconstruction, pp. 79-111 in Cracraft, J. and N. Eldredge (eds.) Phylogenetic Analysis and Paleontology Columbia University Press, New York. Gilmour, J. S. L. (1940) Taxonomy and philosophy, pp. 461-474 in J. Huxley (ed.) The New Systematics Oxford Hull, D. L. (1978) A matter of individuality Phil. of Science 45: 335-360 Hull, D. L. (1978) The principles of biological classification: the use and abuse of philosophy Hull, D. L. (1984) Cladistic theory: hypotheses that blur and grow, pp. 5-23 in T. Duncan and T. F. Stuessy (eds.) Cladistics: Perspectives on the Reconstruction of Evolutionary History Columbia University Press, New York * Hull, D. L. (1988) Science as a process: an evolutionary account of the social and conceptual development of science University of Chicago Press. An already classic work on the recent, violent history of systematics; used as data for Hull's general theories about scientific change. -

Natural Selection and Adaptation, Part I
Natural Selection and Adaptation, Part I 36-149 The Tree of Life Christopher R. Genovese Department of Statistics 132H Baker Hall x8-7836 http://www.stat.cmu.edu/~genovese/ . Plan • Review of Natural Selection • Detecting Natural Selection (discussion) • Examples of Observed Natural Selection ............................... Next Time: • Adaptive Traits • Methods for Reasoning about and studying adaptations • Explaining Complex Adaptations (discussion) 36-149 The Tree of Life Class #10 -1- Overview The theories of common descent and natural selection play different roles within the theory of evolution. Common Descent explains the unity of life. Natural Selection explains the diversity of life. An adaptation (or adaptive trait) is a feature of an organism that enhances reproductive success, relative to other possible variants, in a given environment. Adaptations become prevalent and are maintained in a population through natural selection. Indeed, natural selection is the only mechanism of evolutionary change that can satisfactorily explain adaptations. 36-149 The Tree of Life Class #10 -2- Darwin's Argument Darwin put forward two main arguments in support of natural selection: An analogical argument: Artificial selection A logical argument: The struggle for existence (As we will see later, we now have more than just argument in support of the theory.) 36-149 The Tree of Life Class #10 -3- The Analogical Argument: Artificial Selection 36-149 The Tree of Life Class #10 -4- The Analogical Argument: Artificial Selection 36-149 The Tree of Life Class #10 -5- The Analogical Argument: Artificial Selection Teosinte to Corn 36-149 The Tree of Life Class #10 -6- The Analogical Argument: Artificial Selection • Darwin was intimately familiar with the efforts of breeders in his day to produce novel varieties. -

1. Adaptation and the Evolution of Physiological Characters
Bennett, A. F. 1997. Adaptation and the evolution of physiological characters, pp. 3-16. In: Handbook of Physiology, Sect. 13: Comparative Physiology. W. H. Dantzler, ed. Oxford Univ. Press, New York. 1. Adaptation and the evolution of physiological characters Department of Ecology and Evolutionary Biology, University of California, ALBERT F. BENNETT 1 Irvine, California among the biological sciences (for example, behavioral CHAPTER CONTENTS science [I241). The Many meanings of "Adaptationn In general, comparative physiologists have been Criticisms of Adaptive Interpretations much more successful in, and have devoted much more Alternatives to Adaptive Explanations energy to, pursuing the former rather than the latter Historical inheritance goal (37). Most of this Handbook is devoted to an Developmentai pattern and constraint Physical and biomechanical correlation examination of mechanism-how various physiologi- Phenotypic size correlation cal systems function in various animals. Such compara- Genetic correlations tive studies are usually interpreted within a specific Chance fixation evolutionary context, that of adaptation. That is, or- Studying the Evolution of Physiological Characters ganisms are asserted to be designed in the ways they Macroevolutionary studies Microevolutionary studies are and to function in the ways they do because of Incorporating an Evolutionary Perspective into Physiological Studies natural selection which results in evolutionary change. The principal textbooks in the field (for example, refs. 33, 52, 102, 115) make explicit reference in their titles to the importance of adaptation to comparative COMPARATIVE PHYSIOLOGISTS HAVE TWO GOALS. The physiology, as did the last comparative section of this first is to explain mechanism, the study of how organ- Handbook (32). Adaptive evolutionary explanations isms are built functionally, "how animals work" (113). -

Transcriptional Regulation by Extracellular Signals 209
Cell, Vol. 80, 199-211, January 27, 1995, Copyright © 1995 by Cell Press Transcriptional Regulation Review by Extracellular Signals: Mechanisms and Specificity Caroline S. Hill and Richard Treisman Nuclear Translocation Transcription Laboratory In principle, regulated nuclear localization of transcription Imperial Cancer Research Fund factors can involve regulated activity of either nuclear lo- Lincoln's Inn Fields calization signals (NLSs) or cytoplasmic retention signals, London WC2A 3PX although no well-characterized case of the latter has yet England been reported. N LS activity, which is generally dependent on short regions of basic amino acids, can be regulated either by masking mechanisms or by phosphorylations Changes in cell behavior induced by extracellular signal- within the NLS itself (Hunter and Karin, 1992). For exam- ing molecules such as growth factors and cytokines re- ple, association with an inhibitory subunit masks the NLS quire execution of a complex program of transcriptional of NF-KB and its relatives (Figure 1; for review see Beg events. While the route followed by the intracellular signal and Baldwin, 1993), while an intramolecular mechanism from the cell membrane to its transcription factor targets may mask NLS activity in the heat shock regulatory factor can be traced in an increasing number of cases, how the HSF2 (Sheldon and Kingston, 1993). When transcription specificity of the transcriptional response of the cell to factor localization is dependent on regulated NLS activity, different stimuli is determined is much less clear. How- linkage to a constitutively acting NLS may be sufficient to ever, it is possible to understand at least in principle how render nuclear localization independent of signaling (Beg different stimuli can activate the same signal pathway yet et al., 1992). -

NF-Κb Modifies the Mammalian Circadian Clock Through Interaction with the Core Clock Protein BMAL1
bioRxiv preprint doi: https://doi.org/10.1101/2020.09.06.285254; this version posted September 6, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. NF-κB modifies the mammalian circadian clock through interaction with the core clock protein BMAL1 Yang Shen1, Wei Wang2, Mehari Endale1, Lauren J. Francey3, Rachel L. Harold4, David W. Hammers5, Zhiguang Huo6, Carrie L. Partch4, John B. Hogenesch3, Zhao-Hui Wu2, and Andrew C. Liu1* 1Department of Physiology and Functional Genomics, University of Florida College of Medicine; 2Department of Radiation Oncology and Center for Cancer Research, University of Tennessee Health Science Center; 3Divisions of Human Genetics and Immunobiology, Cincinnati Children's Hospital Medical Center; 4Department of Chemistry and Biochemistry, University of California Santa Cruz; 5Department of Pharmacology and Therapeutics, University of Florida College of Medicine; 6Department of Biostatistics, College of Public Health & Health Professions, University of Florida College of Medicine Running title: NF-κB modifies circadian clock * Correspondence: Andrew C. Liu, [email protected] Keywords: Circadian clock, inflammation, NF-κB, suprachiasmatic nucleus (SCN), BMAL1 Abbreviations: NF-κB, nuclear factor kappa B; SCN, suprachiasmatic nucleus; IKK, IκB kinase; IκBα, inhibitor of NF-κB; Luc, luciferase; RHD, Rel homology domain; CRD, C-terminal oscillation regulatory domain, TAD, transactivation domain 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.09.06.285254; this version posted September 6, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. -

Evolutionary and Historical Biogeography of Animal Diversity Learning Objectives
Evolutionary and historical biogeography of animal diversity Learning objectives • The students can explain the common ancestor of animal kingdom. • The students can explain the historical biogeography of animal. • The students can explain the invasion of animal from aquatic to terrestrial habitat. • The students can explain the basic mechanism of speciation, allopatric and non-allopatric. The Common Ancestor of Animal Kingdom Characteristics of Animals • Animals or “metazoans” are typically heterotrophic, multicellular organisms with diploid, eukaryotic cells. • Trichoplax adhaerens is defined as an animal by the presence of different somatic (i.e., non-reproductive) cell types and by impermeable cell–cell connections. Trichoplax adhaerens Blackstone, 2009 Two Hypotheses for the Branching Order of Groups at the Root of the Metazoan Tree 1 2 The choanoflagellates serve as an outgroup in the Bilaterians are the sister group to the placozoan + analysis, and sponges are the sister group to the sponge + ctenophore + cnidarian clade, while placozoans placozoan + cnidarian + ctenophore + bilaterian are the sister group to the sponge + ctenophore + clade. cnidarian clade. Blackstone, 2009 Ancestry and evolution of animal–bacterial interactions • Choanoflagellates as the last common ancestor of animal kingdom. • Urmetazoan is the group of animal with multicellular and produce differentiated cell types (ex. Egg & sperm) R.A. Alegado & N. King, 2014 Conserved morphology and ultrastructure of Choanoflagellates and Sponge choanocytes The collar complex is conserved in choanoflagellates (A. S. rosetta) and sponge collar cells (B. Sycon coactum) flagellum (fL), microvilli (mv), a nucleus (nu), and a food vacuole (fv) Brunet & King, 2017 The Historical Biogeography of Animal Zoogeographic regions Old New Cox, 2001 Plate tectonic regulation of global marine animal diversity A. -

Auditory Experience Controls the Maturation of Song Discrimination and Sexual Response in Drosophila Xiaodong Li, Hiroshi Ishimoto, Azusa Kamikouchi*
RESEARCH ARTICLE Auditory experience controls the maturation of song discrimination and sexual response in Drosophila Xiaodong Li, Hiroshi Ishimoto, Azusa Kamikouchi* Graduate School of Science, Nagoya University, Nagoya, Japan Abstract In birds and higher mammals, auditory experience during development is critical to discriminate sound patterns in adulthood. However, the neural and molecular nature of this acquired ability remains elusive. In fruit flies, acoustic perception has been thought to be innate. Here we report, surprisingly, that auditory experience of a species-specific courtship song in developing Drosophila shapes adult song perception and resultant sexual behavior. Preferences in the song-response behaviors of both males and females were tuned by social acoustic exposure during development. We examined the molecular and cellular determinants of this social acoustic learning and found that GABA signaling acting on the GABAA receptor Rdl in the pC1 neurons, the integration node for courtship stimuli, regulated auditory tuning and sexual behavior. These findings demonstrate that maturation of auditory perception in flies is unexpectedly plastic and is acquired socially, providing a model to investigate how song learning regulates mating preference in insects. DOI: https://doi.org/10.7554/eLife.34348.001 Introduction Vocal learning in infants or juvenile birds relies heavily on the early experience of the adult conspe- cific sounds. In humans, early language input is necessary to form the ability of phonetic distinction *For correspondence: and pattern detection in the phase of auditory learning (Doupe and Kuhl, 1999; Kuhl, 2004). [email protected] Because of the strong parallels between speech acquisition of humans and song learning of song- Competing interests: The birds, and the difficulties to investigate the neural mechanisms of human early auditory memory at authors declare that no cellular resolution, songbirds have been used as a predominant model in studying memory formation competing interests exist. -
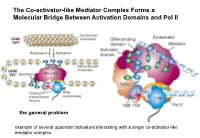
The Co-Activator-Like Mediator Complex Forms a Molecular Bridge Between Activation Domains and Pol II
The Co-activator-like Mediator Complex Forms a Molecular Bridge Between Activation Domains and Pol II Co-activator the general problem example of several upstream activators interacting with a single co-activator-like mediator complex Transcription Regulation And Gene Expression in Eukaryotes FS 2016 Graduate Course G2 P. Matthias and RG Clerc Pharmazentrum Hörsaal 2 16h15-18h00 REGULATORY MECHANISMS OF TRANSCRIPTION FACTOR FUNCTION •Protein synthesized •Protein phosphorylated •Ligand binding •Release inhibitor •Change partner, etc RG Clerc April 6. 2016 Transcription factors as final effectors of the cellular signaling cascade Regulatory Mechanisms of Transcription Factor Function Genes X. Lewin B. editor Regulatory Mechanisms of Transcription Factor Function CREB FOXO NFAT Genes X. Lewin B. editor Body plan is constructed through interactions of the developmentally regulated homeotic gene expression anterior early expression posterior late expression high RA response low RA response hindbrain trunk Alexander T and Krumlauf R. Ann.Rev.Cell.Dev.Biol: 25_431 (2009) Hoxb transcription factors mRNA distribution along the AP axis A staggered expression of the anterior border within somites is a property of the physical ordering along the chromosome: the colinearity Alexander T and Krumlauf R. Ann.Rev.Cell.Dev.Biol: 25_431 (2009) Transcription control of the Hox genes: insight into colinear activation P A p p p p a p a p a a a a Tarchini B and Duboule D. Dev.Cell 10_93 (2006) correlation between linear arrangement along the chromosome and spatial -

The C-Rel Transcription Factor and B-Cell Proliferation: a Deal with the Devil
Oncogene (2004) 23, 2275–2286 & 2004 Nature Publishing Group All rights reserved 0950-9232/04 $25.00 www.nature.com/onc REVIEW The c-Rel transcription factor and B-cell proliferation: a deal with the devil Thomas D Gilmore*,1, Demetrios Kalaitzidis1, Mei-Chih Liang1 and Daniel T Starczynowski1 1Department of Biology, Boston University, 5 Cummington Street, Boston, MA 02215, USA Activation of the Rel/NF-jB signal transduction pathway called the Rel homology domain, which has sequences has been associated with a varietyof animal and human required for DNA binding, dimerization, nuclear malignancies. However, among the Rel/NF-jB family localization, and inhibitor (IkB) binding. Sequences members, onlyc-Rel has been consistentlyshown to be C-terminal to the Rel homology domain in c-Rel, RelA, able to malignantlytransform cells in culture. In addition, and RelB contain transcriptional activation domains, c-rel has been activated bya retroviral promoter insertion and therefore dimers that contain these Rel/NF-kB in an avian B-cell lymphoma, and amplifications of REL members usually increase transcription of target genes. (human c-rel) are frequentlyseen in Hodgkin’s lympho- Although the Rel homology domain sequences of c-Rel mas and diffuse large B-cell lymphomas, and in some proteins across species are highly conserved, their follicular and mediastinal B-cell lymphomas. Phenotypic C-terminal transactivation domains are not. For exam- analysis of c-rel knockout mice demonstrates that c-Rel ple, the Rel homology domains of chicken and human has a normal role in B-cell proliferation and survival; c-Rel are approximately 85% identical, whereas their moreover, c-Rel nuclear activityis required for B-cell C-terminal transactivation domains are only about 10% development. -

Reducing Risks Through Adaptation Actions | Fourth National Climate
Impacts, Risks, and Adaptation in the United States: Fourth National Climate Assessment, Volume II 28 Reducing Risks Through Adaptation Actions Federal Coordinating Lead Authors Review Editor Jeffrey Arnold Mary Ann Lazarus U.S. Army Corps of Engineers Cameron MacAllister Group Roger Pulwarty National Oceanic and Atmospheric Administration Chapter Lead Robert Lempert RAND Corporation Chapter Authors Kate Gordon Paulson Institute Katherine Greig Wharton Risk Management and Decision Processes Center at University of Pennsylvania (formerly New York City Mayor’s Office of Recovery and Resiliency) Cat Hawkins Hoffman National Park Service Dale Sands Village of Deer Park, Illinois Caitlin Werrell The Center for Climate and Security Technical Contributors are listed at the end of the chapter. Recommended Citation for Chapter Lempert, R., J. Arnold, R. Pulwarty, K. Gordon, K. Greig, C. Hawkins Hoffman, D. Sands, and C. Werrell, 2018: Reducing Risks Through Adaptation Actions. In Impacts, Risks, and Adaptation in the United States: Fourth National Climate Assessment, Volume II [Reidmiller, D.R., C.W. Avery, D.R. Easterling, K.E. Kunkel, K.L.M. Lewis, T.K. Maycock, and B.C. Stewart (eds.)]. U.S. Global Change Research Program, Washington, DC, USA, pp. 1309–1345. doi: 10.7930/NCA4.2018.CH28 On the Web: https://nca2018.globalchange.gov/chapter/adaptation Impacts, Risks, and Adaptation in the United States: Fourth National Climate Assessment, Volume II 28 Reducing Risks Through Adaptation Actions Key Message 1 Seawall surrounding Kivalina, Alaska Adaptation Implementation Is Increasing Adaptation planning and implementation activities are occurring across the United States in the public, private, and nonprofit sectors. Since the Third National Climate Assessment, implementation has increased but is not yet commonplace. -

Protist Phylogeny and the High-Level Classification of Protozoa
Europ. J. Protistol. 39, 338–348 (2003) © Urban & Fischer Verlag http://www.urbanfischer.de/journals/ejp Protist phylogeny and the high-level classification of Protozoa Thomas Cavalier-Smith Department of Zoology, University of Oxford, South Parks Road, Oxford, OX1 3PS, UK; E-mail: [email protected] Received 1 September 2003; 29 September 2003. Accepted: 29 September 2003 Protist large-scale phylogeny is briefly reviewed and a revised higher classification of the kingdom Pro- tozoa into 11 phyla presented. Complementary gene fusions reveal a fundamental bifurcation among eu- karyotes between two major clades: the ancestrally uniciliate (often unicentriolar) unikonts and the an- cestrally biciliate bikonts, which undergo ciliary transformation by converting a younger anterior cilium into a dissimilar older posterior cilium. Unikonts comprise the ancestrally unikont protozoan phylum Amoebozoa and the opisthokonts (kingdom Animalia, phylum Choanozoa, their sisters or ancestors; and kingdom Fungi). They share a derived triple-gene fusion, absent from bikonts. Bikonts contrastingly share a derived gene fusion between dihydrofolate reductase and thymidylate synthase and include plants and all other protists, comprising the protozoan infrakingdoms Rhizaria [phyla Cercozoa and Re- taria (Radiozoa, Foraminifera)] and Excavata (phyla Loukozoa, Metamonada, Euglenozoa, Percolozoa), plus the kingdom Plantae [Viridaeplantae, Rhodophyta (sisters); Glaucophyta], the chromalveolate clade, and the protozoan phylum Apusozoa (Thecomonadea, Diphylleida). Chromalveolates comprise kingdom Chromista (Cryptista, Heterokonta, Haptophyta) and the protozoan infrakingdom Alveolata [phyla Cilio- phora and Miozoa (= Protalveolata, Dinozoa, Apicomplexa)], which diverged from a common ancestor that enslaved a red alga and evolved novel plastid protein-targeting machinery via the host rough ER and the enslaved algal plasma membrane (periplastid membrane). -

The Evolution of Bird Song: Male and Female Response to Song Innovation in Swamp Sparrows
ANIMAL BEHAVIOUR, 2001, 62, 1189–1195 doi:10.1006/anbe.2001.1854, available online at http://www.idealibrary.com on The evolution of bird song: male and female response to song innovation in swamp sparrows STEPHEN NOWICKI*, WILLIAM A. SEARCY†, MELISSA HUGHES‡ & JEFFREY PODOS§ *Evolution, Ecology & Organismal Biology Group, Department of Biology, Duke University †Department of Biology, University of Miami ‡Department of Ecology and Evolutionary Biology, Princeton University §Department of Biology, University of Massachusetts at Amherst (Received 27 April 2000; initial acceptance 24 July 2000; final acceptance 19 April 2001; MS. number: A8776) Closely related species of songbirds often show large differences in song syntax, suggesting that major innovations in syntax must sometimes arise and spread. Here we examine the response of male and female swamp sparrows, Melospiza georgiana, to an innovation in song syntax produced by males of this species. Young male swamp sparrows that have been exposed to tutor songs with experimentally increased trill rates reproduce these songs with periodic silent gaps (Podos 1996, Animal Behaviour, 51, 1061–1070). This novel temporal pattern, termed ‘broken syntax’, has been demonstrated to transmit across generations (Podos et al. 1999, Animal Behaviour, 58, 93–103). We show here that adult male swamp sparrows respond more strongly in territorial playback tests to songs with broken syntax than to heterospecific songs, and equally strongly to conspecific songs with normal and broken syntax. In tests using the solicitation display assay, adult female swamp sparrows respond more to broken syntax than to heterospecific songs, although they respond significantly less to conspecific songs with broken syntax than to those with normal syntax.