Full Text in Pdf Format
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Selecting Molecular Markers for a Specific Phylogenetic Problem
MOJ Proteomics & Bioinformatics Review Article Open Access Selecting molecular markers for a specific phylogenetic problem Abstract Volume 6 Issue 3 - 2017 In a molecular phylogenetic analysis, different markers may yield contradictory Claudia AM Russo, Bárbara Aguiar, Alexandre topologies for the same diversity group. Therefore, it is important to select suitable markers for a reliable topological estimate. Issues such as length and rate of evolution P Selvatti Department of Genetics, Federal University of Rio de Janeiro, will play a role in the suitability of a particular molecular marker to unfold the Brazil phylogenetic relationships for a given set of taxa. In this review, we provide guidelines that will be useful to newcomers to the field of molecular phylogenetics weighing the Correspondence: Claudia AM Russo, Molecular Biodiversity suitability of molecular markers for a given phylogenetic problem. Laboratory, Department of Genetics, Institute of Biology, Block A, CCS, Federal University of Rio de Janeiro, Fundão Island, Keywords: phylogenetic trees, guideline, suitable genes, phylogenetics Rio de Janeiro, RJ, 21941-590, Brazil, Tel 21 991042148, Fax 21 39386397, Email [email protected] Received: March 14, 2017 | Published: November 03, 2017 Introduction concept that occupies a central position in evolutionary biology.15 Homology is a qualitative term, defined by equivalence of parts due Over the last three decades, the scientific field of molecular to inherited common origin.16–18 biology has experienced remarkable advancements -

An Automated Pipeline for Retrieving Orthologous DNA Sequences from Genbank in R
life Technical Note phylotaR: An Automated Pipeline for Retrieving Orthologous DNA Sequences from GenBank in R Dominic J. Bennett 1,2,* ID , Hannes Hettling 3, Daniele Silvestro 1,2, Alexander Zizka 1,2, Christine D. Bacon 1,2, Søren Faurby 1,2, Rutger A. Vos 3 ID and Alexandre Antonelli 1,2,4,5 ID 1 Gothenburg Global Biodiversity Centre, Box 461, SE-405 30 Gothenburg, Sweden; [email protected] (D.S.); [email protected] (A.Z.); [email protected] (C.D.B.); [email protected] (S.F.); [email protected] (A.A.) 2 Department of Biological and Environmental Sciences, University of Gothenburg, Box 461, SE-405 30 Gothenburg, Sweden 3 Naturalis Biodiversity Center, P.O. Box 9517, 2300 RA Leiden, The Netherlands; [email protected] (H.H.); [email protected] (R.A.V.) 4 Gothenburg Botanical Garden, Carl Skottsbergsgata 22A, SE-413 19 Gothenburg, Sweden 5 Department of Organismic and Evolutionary Biology, Harvard University, 26 Oxford St., Cambridge, MA 02138 USA * Correspondence: [email protected] Received: 28 March 2018; Accepted: 1 June 2018; Published: 5 June 2018 Abstract: The exceptional increase in molecular DNA sequence data in open repositories is mirrored by an ever-growing interest among evolutionary biologists to harvest and use those data for phylogenetic inference. Many quality issues, however, are known and the sheer amount and complexity of data available can pose considerable barriers to their usefulness. A key issue in this domain is the high frequency of sequence mislabeling encountered when searching for suitable sequences for phylogenetic analysis. -

Fishery Management Plan for Groundfish of the Bering Sea and Aleutian Islands Management Area APPENDICES
FMP for Groundfish of the BSAI Management Area Fishery Management Plan for Groundfish of the Bering Sea and Aleutian Islands Management Area APPENDICES Appendix A History of the Fishery Management Plan ...................................................................... A-1 A.1 Amendments to the FMP ......................................................................................................... A-1 Appendix B Geographical Coordinates of Areas Described in the Fishery Management Plan ..... B-1 B.1 Management Area, Subareas, and Districts ............................................................................. B-1 B.2 Closed Areas ............................................................................................................................ B-2 B.3 PSC Limitation Zones ........................................................................................................... B-18 Appendix C Summary of the American Fisheries Act and Subtitle II ............................................. C-1 C.1 Summary of the American Fisheries Act (AFA) Management Measures ............................... C-1 C.2 Summary of Amendments to AFA in the Coast Guard Authorization Act of 2010 ................ C-2 C.3 American Fisheries Act: Subtitle II Bering Sea Pollock Fishery ............................................ C-4 Appendix D Life History Features and Habitat Requirements of Fishery Management Plan SpeciesD-1 D.1 Walleye pollock (Theragra calcogramma) ............................................................................ -

"Phylogenetic Analysis of Protein Sequence Data Using The
Phylogenetic Analysis of Protein Sequence UNIT 19.11 Data Using the Randomized Axelerated Maximum Likelihood (RAXML) Program Antonis Rokas1 1Department of Biological Sciences, Vanderbilt University, Nashville, Tennessee ABSTRACT Phylogenetic analysis is the study of evolutionary relationships among molecules, phenotypes, and organisms. In the context of protein sequence data, phylogenetic analysis is one of the cornerstones of comparative sequence analysis and has many applications in the study of protein evolution and function. This unit provides a brief review of the principles of phylogenetic analysis and describes several different standard phylogenetic analyses of protein sequence data using the RAXML (Randomized Axelerated Maximum Likelihood) Program. Curr. Protoc. Mol. Biol. 96:19.11.1-19.11.14. C 2011 by John Wiley & Sons, Inc. Keywords: molecular evolution r bootstrap r multiple sequence alignment r amino acid substitution matrix r evolutionary relationship r systematics INTRODUCTION the baboon-colobus monkey lineage almost Phylogenetic analysis is a standard and es- 25 million years ago, whereas baboons and sential tool in any molecular biologist’s bioin- colobus monkeys diverged less than 15 mil- formatics toolkit that, in the context of pro- lion years ago (Sterner et al., 2006). Clearly, tein sequence analysis, enables us to study degree of sequence similarity does not equate the evolutionary history and change of pro- with degree of evolutionary relationship. teins and their function. Such analysis is es- A typical phylogenetic analysis of protein sential to understanding major evolutionary sequence data involves five distinct steps: (a) questions, such as the origins and history of data collection, (b) inference of homology, (c) macromolecules, developmental mechanisms, sequence alignment, (d) alignment trimming, phenotypes, and life itself. -

New Syllabus
M.Sc. BIOINFORMATICS REGULATIONS AND SYLLABI (Effective from 2019-2020) Centre for Bioinformatics SCHOOL OF LIFE SCIENCES PONDICHERRY UNIVERSITY PUDUCHERRY 1 Pondicherry University School of Life Sciences Centre for Bioinformatics Master of Science in Bioinformatics The M.Sc Bioinformatics course started since 2007 under UGC Innovative program Program Objectives The main objective of the program is to train the students to learn an innovative and evolving field of bioinformatics with a multi-disciplinary approach. Hands-on sessions will be provided to train the students in both computer and experimental labs. Program Outcomes On completion of this program, students will be able to: Gain understanding of the principles and concepts of both biology along with computer science To use and describe bioinformatics data, information resource and also to use the software effectively from large databases To know how bioinformatics methods can be used to relate sequence to structure and function To develop problem-solving skills, new algorithms and analysis methods are learned to address a range of biological questions. 2 Eligibility for M.Sc. Bioinformatics Students from any of the below listed Bachelor degrees with minimum 55% of marks are eligible. Bachelor’s degree in any relevant area of Physics / Chemistry / Computer Science / Life Science with a minimum of 55% of marks 3 PONDICHERRY UNIVERSITY SCHOOL OF LIFE SCIENCES CENTRE FOR BIOINFORMATICS LIST OF HARD-CORE COURSES FOR M.Sc. BIOINFORMATICS (Academic Year 2019-2020 onwards) Course -

Historical Fish Specimens Collected from the Tohoku District by the Saito Ho-On Kai Museum of Natural History
Bull. Natl. Mus. Nat. Sci., Ser. A, 35(1), pp. 9–54, March 22, 2009 Historical Fish Specimens Collected from the Tohoku District by the Saito Ho-on Kai Museum of Natural History Keiichi Matsuura1, Gento Shinohara2 and Masanori Nakae1 1 Collection Center, National Museum of Nature and Science, 3–23–1 Hyakunin-cho, Shinjuku-ku, Tokyo, 169–0073 Japan E-mail: [email protected]; [email protected] 2 Department of Zoology, National Museum of Nature and Science, 3–23–1 Hyakunin-cho, Shinjuku-ku, Tokyo, 169–0073 Japan E-mail: [email protected] Abstract The fish collection of the Saito Ho-on Kai Museum of Natural History was transferred to the National Museum of Nature and Science, Tokyo in February 2006. Ninety percent of the fish collection contains specimens collected from the Tohoku District during the period from 1930 to 1933 when natural environments of Japan were in good condition for various groups of fishes. The fish specimens from the Tohoku District were classified into 361 species/subspecies of 273 genera belonging to 131 families of 31 orders. A list of the species is shown with remarks on distribution. Key words: Fish specimens, Saito Ho-on Kai Museum, Tohoku District, inventory. stead of natural sicence. The museum has tried to Introduction keep its activity at the level before the war, but it The Saito Ho-on Kai Museum was established failed to do so because of financial difficulties. In in November 1933 in Sendai City, Miyagi Pre- 2005, the Saito Ho-on Kai Museum of Natural fecture, Japan. -

Molecular Phylogenetics Suggests a New Classification and Uncovers Convergent Evolution of Lithistid Demosponges
RESEARCH ARTICLE Deceptive Desmas: Molecular Phylogenetics Suggests a New Classification and Uncovers Convergent Evolution of Lithistid Demosponges Astrid Schuster1,2, Dirk Erpenbeck1,3, Andrzej Pisera4, John Hooper5,6, Monika Bryce5,7, Jane Fromont7, Gert Wo¨ rheide1,2,3* 1. Department of Earth- & Environmental Sciences, Palaeontology and Geobiology, Ludwig-Maximilians- Universita¨tMu¨nchen, Richard-Wagner Str. 10, 80333 Munich, Germany, 2. SNSB – Bavarian State Collections OPEN ACCESS of Palaeontology and Geology, Richard-Wagner Str. 10, 80333 Munich, Germany, 3. GeoBio-CenterLMU, Ludwig-Maximilians-Universita¨t Mu¨nchen, Richard-Wagner Str. 10, 80333 Munich, Germany, 4. Institute of Citation: Schuster A, Erpenbeck D, Pisera A, Paleobiology, Polish Academy of Sciences, ul. Twarda 51/55, 00-818 Warszawa, Poland, 5. Queensland Hooper J, Bryce M, et al. (2015) Deceptive Museum, PO Box 3300, South Brisbane, QLD 4101, Australia, 6. Eskitis Institute for Drug Discovery, Griffith Desmas: Molecular Phylogenetics Suggests a New Classification and Uncovers Convergent Evolution University, Nathan, QLD 4111, Australia, 7. Department of Aquatic Zoology, Western Australian Museum, of Lithistid Demosponges. PLoS ONE 10(1): Locked Bag 49, Welshpool DC, Western Australia, 6986, Australia e116038. doi:10.1371/journal.pone.0116038 *[email protected] Editor: Mikhail V. Matz, University of Texas, United States of America Received: July 3, 2014 Accepted: November 30, 2014 Abstract Published: January 7, 2015 Reconciling the fossil record with molecular phylogenies to enhance the Copyright: ß 2015 Schuster et al. This is an understanding of animal evolution is a challenging task, especially for taxa with a open-access article distributed under the terms of the Creative Commons Attribution License, which mostly poor fossil record, such as sponges (Porifera). -

A New Species of Torrent Catfish, Liobagrus Hyeongsanensis (Teleostei: Siluriformes: Amblycipitidae), from Korea
Zootaxa 4007 (1): 267–275 ISSN 1175-5326 (print edition) www.mapress.com/zootaxa/ Article ZOOTAXA Copyright © 2015 Magnolia Press ISSN 1175-5334 (online edition) http://dx.doi.org/10.11646/zootaxa.4007.2.9 http://zoobank.org/urn:lsid:zoobank.org:pub:60ABECAF-9687-4172-A309-D2222DFEC473 A new species of torrent catfish, Liobagrus hyeongsanensis (Teleostei: Siluriformes: Amblycipitidae), from Korea SU-HWAN KIM1, HYEONG-SU KIM2 & JONG-YOUNG PARK2,3 1National Institute of Ecology, Seocheon 325-813, South Korea 2Department of Biological Sciences, College of Natural Sciences, and Institute for Biodiversity Research, Chonbuk National Univer- sity, Jeonju 561-756, South Korea 3Corresponding author. E-mail: [email protected] Abstract A new species of torrent catfish, Liobargus hyeongsanensis, is described from rivers and tributaries of the southeastern coast of Korea. The new species can be differentiated from its congeners by the following characteristics: a small size with a maximum standard length (SL) of 90 mm; body and fins entirely brownish-yellow without distinct markings; a relatively short pectoral spine (3.7–6.5 % SL); a reduced body-width at pectoral-fin base (15.5–17.9 % SL); 50–54 caudal-fin rays; 6–8 gill rakers; 2–3 (mostly 3) serrations on pectoral fin; 60–110 eggs per gravid female. Key words: Amblycipitidae, Liobagrus hyeongsanensis, New species, Endemic, South Korea Introduction Species of the family Amblycipitidae, which comprises four genera, are found in swift freshwater streams in southern and eastern Asia, ranging from Pakistan across northern India to Malaysia, Korea, and Southern Japan (Chen & Lundberg 1995; Ng & Kottelat 2000; Kim & Park 2002; Wright & Ng 2008). -

Humboldt Bay Fishes
Humboldt Bay Fishes ><((((º>`·._ .·´¯`·. _ .·´¯`·. ><((((º> ·´¯`·._.·´¯`·.. ><((((º>`·._ .·´¯`·. _ .·´¯`·. ><((((º> Acknowledgements The Humboldt Bay Harbor District would like to offer our sincere thanks and appreciation to the authors and photographers who have allowed us to use their work in this report. Photography and Illustrations We would like to thank the photographers and illustrators who have so graciously donated the use of their images for this publication. Andrey Dolgor Dan Gotshall Polar Research Institute of Marine Sea Challengers, Inc. Fisheries And Oceanography [email protected] [email protected] Michael Lanboeuf Milton Love [email protected] Marine Science Institute [email protected] Stephen Metherell Jacques Moreau [email protected] [email protected] Bernd Ueberschaer Clinton Bauder [email protected] [email protected] Fish descriptions contained in this report are from: Froese, R. and Pauly, D. Editors. 2003 FishBase. Worldwide Web electronic publication. http://www.fishbase.org/ 13 August 2003 Photographer Fish Photographer Bauder, Clinton wolf-eel Gotshall, Daniel W scalyhead sculpin Bauder, Clinton blackeye goby Gotshall, Daniel W speckled sanddab Bauder, Clinton spotted cusk-eel Gotshall, Daniel W. bocaccio Bauder, Clinton tube-snout Gotshall, Daniel W. brown rockfish Gotshall, Daniel W. yellowtail rockfish Flescher, Don american shad Gotshall, Daniel W. dover sole Flescher, Don stripped bass Gotshall, Daniel W. pacific sanddab Gotshall, Daniel W. kelp greenling Garcia-Franco, Mauricio louvar -
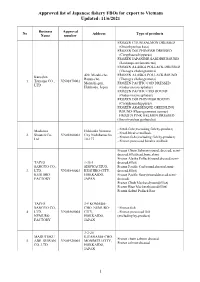
Approved List of Japanese Fishery Fbos for Export to Vietnam Updated: 11/6/2021
Approved list of Japanese fishery FBOs for export to Vietnam Updated: 11/6/2021 Business Approval No Address Type of products Name number FROZEN CHUM SALMON DRESSED (Oncorhynchus keta) FROZEN DOLPHINFISH DRESSED (Coryphaena hippurus) FROZEN JAPANESE SARDINE ROUND (Sardinops melanostictus) FROZEN ALASKA POLLACK DRESSED (Theragra chalcogramma) 420, Misaki-cho, FROZEN ALASKA POLLACK ROUND Kaneshin Rausu-cho, (Theragra chalcogramma) 1. Tsuyama CO., VN01870001 Menashi-gun, FROZEN PACIFIC COD DRESSED LTD Hokkaido, Japan (Gadus macrocephalus) FROZEN PACIFIC COD ROUND (Gadus macrocephalus) FROZEN DOLPHIN FISH ROUND (Coryphaena hippurus) FROZEN ARABESQUE GREENLING ROUND (Pleurogrammus azonus) FROZEN PINK SALMON DRESSED (Oncorhynchus gorbuscha) - Fresh fish (excluding fish by-product) Maekawa Hokkaido Nemuro - Fresh bivalve mollusk. 2. Shouten Co., VN01860002 City Nishihamacho - Frozen fish (excluding fish by-product) Ltd 10-177 - Frozen processed bivalve mollusk Frozen Chum Salmon (round, dressed, semi- dressed,fillet,head,bone,skin) Frozen Alaska Pollack(round,dressed,semi- TAIYO 1-35-1 dressed,fillet) SANGYO CO., SHOWACHUO, Frozen Pacific Cod(round,dressed,semi- 3. LTD. VN01840003 KUSHIRO-CITY, dressed,fillet) KUSHIRO HOKKAIDO, Frozen Pacific Saury(round,dressed,semi- FACTORY JAPAN dressed) Frozen Chub Mackerel(round,fillet) Frozen Blue Mackerel(round,fillet) Frozen Salted Pollack Roe TAIYO 3-9 KOMABA- SANGYO CO., CHO, NEMURO- - Frozen fish 4. LTD. VN01860004 CITY, - Frozen processed fish NEMURO HOKKAIDO, (excluding by-product) FACTORY JAPAN -

Fishes-Of-The-Salish-Sea-Pp18.Pdf
NOAA Professional Paper NMFS 18 Fishes of the Salish Sea: a compilation and distributional analysis Theodore W. Pietsch James W. Orr September 2015 U.S. Department of Commerce NOAA Professional Penny Pritzker Secretary of Commerce Papers NMFS National Oceanic and Atmospheric Administration Kathryn D. Sullivan Scientifi c Editor Administrator Richard Langton National Marine Fisheries Service National Marine Northeast Fisheries Science Center Fisheries Service Maine Field Station Eileen Sobeck 17 Godfrey Drive, Suite 1 Assistant Administrator Orono, Maine 04473 for Fisheries Associate Editor Kathryn Dennis National Marine Fisheries Service Offi ce of Science and Technology Fisheries Research and Monitoring Division 1845 Wasp Blvd., Bldg. 178 Honolulu, Hawaii 96818 Managing Editor Shelley Arenas National Marine Fisheries Service Scientifi c Publications Offi ce 7600 Sand Point Way NE Seattle, Washington 98115 Editorial Committee Ann C. Matarese National Marine Fisheries Service James W. Orr National Marine Fisheries Service - The NOAA Professional Paper NMFS (ISSN 1931-4590) series is published by the Scientifi c Publications Offi ce, National Marine Fisheries Service, The NOAA Professional Paper NMFS series carries peer-reviewed, lengthy original NOAA, 7600 Sand Point Way NE, research reports, taxonomic keys, species synopses, fl ora and fauna studies, and data- Seattle, WA 98115. intensive reports on investigations in fi shery science, engineering, and economics. The Secretary of Commerce has Copies of the NOAA Professional Paper NMFS series are available free in limited determined that the publication of numbers to government agencies, both federal and state. They are also available in this series is necessary in the transac- exchange for other scientifi c and technical publications in the marine sciences. -

Independent Evolution of the Specialized Pharyngeal Jaw Apparatus in Cichlid and Labrid Fishes Kohji Mabuchi*1, Masaki Miya2, Yoichiro Azuma1 and Mutsumi Nishida1
BMC Evolutionary Biology BioMed Central Research article Open Access Independent evolution of the specialized pharyngeal jaw apparatus in cichlid and labrid fishes Kohji Mabuchi*1, Masaki Miya2, Yoichiro Azuma1 and Mutsumi Nishida1 Address: 1Ocean Research Institute, The University of Tokyo, 1-15-1 Minamidai, Nakano-ku, Tokyo 164-8639, Japan and 2Department of Zoology, Natural History Museum & Institute, Chiba, 955-2 Aoba-cho, Chuo-ku, Chiba 260-8682, Japan Email: Kohji Mabuchi* - [email protected]; Masaki Miya - [email protected]; Yoichiro Azuma - [email protected]; Mutsumi Nishida - [email protected] * Corresponding author Published: 30 January 2007 Received: 1 August 2006 Accepted: 30 January 2007 BMC Evolutionary Biology 2007, 7:10 doi:10.1186/1471-2148-7-10 This article is available from: http://www.biomedcentral.com/1471-2148/7/10 © 2007 Mabuchi et al; licensee BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Abstract Background: Fishes in the families Cichlidae and Labridae provide good probable examples of vertebrate adaptive radiations. Their spectacular trophic radiations have been widely assumed to be due to structural key innovation in pharyngeal jaw apparatus (PJA), but this idea has never been tested based on a reliable phylogeny. For the first step of evaluating the hypothesis, we investigated the phylogenetic positions of the components of the suborder Labroidei (including Pomacentridae and Embiotocidae in addition to Cichlidae and Labridae) within the Percomorpha, the most diversified (> 15,000 spp) crown clade of teleosts.