Tracking Character Evolution and Biogeographic History Through Time in Cornaceae—Does Choice of Methods Matter? Qiu-Yun (Jenny) XIANG* David T
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
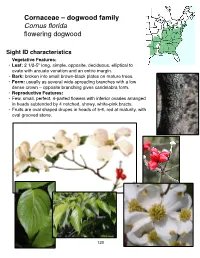
Cornaceae – Dogwood Family Cornus Florida Flowering Dogwood
Cornaceae – dogwood family Cornus florida flowering dogwood Sight ID characteristics Vegetative Features: • Leaf: 2 1/2-5" long, simple, opposite, deciduous, elliptical to ovate with arcuate venation and an entire margin. • Bark: broken into small brown-black plates on mature trees. • Form: usually as several wide-spreading branches with a low dense crown – opposite branching gives candelabra form. • Reproductive Features: • Few, small, perfect, 4-parted flowers with inferior ovaries arranged in heads subtended by 4 notched, showy, white-pink bracts. • Fruits are oval shaped drupes in heads of 5-6, red at maturity, with oval grooved stone. 123 NOTES AND SKETCHES 124 Cornaceae – dogwood family Cornus nuttallii Pacific dogwood Sight ID characteristics Vegetative Features: • Leaf: 2 1/2-4 1/2" long, simple, opposite, deciduous, ovate- elliptical with arcuate venation, margin may be sparsely toothed or entire. • Bark: dark and broken into small plates at maturity. • Form: straight trunk and narrow crown in forested conditions, many-trunked and bushy in open. • Reproductive Features: • Many yellowish-green, small, perfect, 4-parted flowers with inferior ovaries arranged in dense in heads, subtended by 4-7 showy white- pink, petal-like bracts - not notched at the apex. • Fruits are drupes in heads of 30-40, red at maturity and they have smooth stones. 125 NOTES AND SKETCHES 126 Cornaceae – dogwood family Cornus sericea red-osier dogwood Sight ID characteristics Vegetative Features: • Leaf: 2-4" long, simple, opposite, deciduous and somewhat narrow ovate-lanceolate with entire margin. • Twig: bright red, sometimes green splotched with red, white pith. • Bark: red to green with numerous lenticels; later developing larger cracks and splits and turning light brown. -

Department of Planning and Zoning
Department of Planning and Zoning Subject: Howard County Landscape Manual Updates: Recommended Street Tree List (Appendix B) and Recommended Plant List (Appendix C) - Effective July 1, 2010 To: DLD Review Staff Homebuilders Committee From: Kent Sheubrooks, Acting Chief Division of Land Development Date: July 1, 2010 Purpose: The purpose of this policy memorandum is to update the Recommended Plant Lists presently contained in the Landscape Manual. The plant lists were created for the first edition of the Manual in 1993 before information was available about invasive qualities of certain recommended plants contained in those lists (Norway Maple, Bradford Pear, etc.). Additionally, diseases and pests have made some other plants undesirable (Ash, Austrian Pine, etc.). The Howard County General Plan 2000 and subsequent environmental and community planning publications such as the Route 1 and Route 40 Manuals and the Green Neighborhood Design Guidelines have promoted the desirability of using native plants in landscape plantings. Therefore, this policy seeks to update the Recommended Plant Lists by identifying invasive plant species and disease or pest ridden plants for their removal and prohibition from further planting in Howard County and to add other available native plants which have desirable characteristics for street tree or general landscape use for inclusion on the Recommended Plant Lists. Please note that a comprehensive review of the street tree and landscape tree lists were conducted for the purpose of this update, however, only -

The Pollen of Genus Alangium in Cenozoic Deposits of Georgia
saqarTvelos mecnierebaTa erovnuli akademiis moambe, t. 8, #3, 2014 BULLETIN OF THE GEORGIAN NATIONAL ACADEMY OF SCIENCES, vol. 8, no. 3, 2014 Palaeobiology The Pollen of Genus Alangium in Cenozoic Deposits of Georgia Irina Shatilova*, Irma Kokolashvili** * Georgian National Museum, Institute of Palaeobiology, Tbilisi **Georgian Technical University, Tbilisi (Presented by Academy Member David Lordkipanidze) ABSTRACT. In geological records the genus Alangium is known from the Paleogene. Morphologically different pollen grains were determined in the Eocene deposits of both hemispheres: the pollen of Alangium sp. A in North America, Alangiopollis eocaenicus Krutzsch in Europe and Alangium sibiricum Lubomirova in the Western Siberia. The species A. barhoornianum Traverse was described from the Upper Oligocene Brandon lignite of Vermond (North America). The same pollen grains were revealed in Europe. The European pollen remains were similar to Traverse’s Alangium barhoornianum and only the generic name was changed. On the territory of Europe the species Alangiopollis barhoornianum (Traverse) Krutzch is known mainly from the Paleogene till the Middle Miocene. Approximately in the same interval of time the species A. simplex Nagy and A. rarus Cernjavska were determined. On the territory of Georgia the genus Alangium is also known from Paleogene. The species Alangiopollis eocaenicus was described from the Middle Oligocene deposits of Southern Georgia. In Sarmatian the genus was represented by two species A. eocaenicus and A. barhoornianum. After Sarmatian the history of Alangium was connected only with the area adjoining the Black Sea, where the accumulation of marine deposits continued during the whole Pliocene and Pleistocene. In Meotian the pollen grains of two species were determined - the extinct taxon A. -

Caterpillars Moths Butterflies Woodies
NATIVE Caterpillars Moths and utter flies Band host NATIVE Hackberry Emperor oodies PHOTO : Megan McCarty W Double-toothed Prominent Honey locust Moth caterpillar Hackberry Emperor larva PHOTO : Douglas Tallamy Big Poplar Sphinx Number of species of Caterpillars n a study published in 2009, Dr. Oaks (Quercus) 557 Beeches (Fagus) 127 Honey-locusts (Gleditsia) 46 Magnolias (Magnolia) 21 Double-toothed Prominent ( Nerice IDouglas W. Tallamy, Ph.D, chair of the Cherries (Prunus) 456 Serviceberry (Amelanchier) 124 New Jersey Tea (Ceanothus) 45 Buttonbush (Cephalanthus) 19 bidentata ) larvae feed exclusively on elms Department of Entomology and Wildlife Willows (Salix) 455 Larches or Tamaracks (Larix) 121 Sycamores (Platanus) 45 Redbuds (Cercis) 19 (Ulmus), and can be found June through Ecology at the University of Delaware Birches (Betula) 411 Dogwoods (Cornus) 118 Huckleberry (Gaylussacia) 44 Green-briar (Smilax) 19 October. Their body shape mimics the specifically addressed the usefulness of Poplars (Populus) 367 Firs (Abies) 117 Hackberry (Celtis) 43 Wisterias (Wisteria) 19 toothed shape of American elm, making native woodies as host plants for our Crabapples (Malus) 308 Bayberries (Myrica) 108 Junipers (Juniperus) 42 Redbay (native) (Persea) 18 them hard to spot. The adult moth is native caterpillars (and obviously Maples (Acer) 297 Viburnums (Viburnum) 104 Elders (Sambucus) 42 Bearberry (Arctostaphylos) 17 small with a wingspan of 3-4 cm. therefore moths and butterflies). Blueberries (Vaccinium) 294 Currants (Ribes) 99 Ninebark (Physocarpus) 41 Bald cypresses (Taxodium) 16 We present here a partial list, and the Alders (Alnus) 255 Hop Hornbeam (Ostrya) 94 Lilacs (Syringa) 40 Leatherleaf (Chamaedaphne) 15 Honey locust caterpillar feeds on honey number of Lepidopteran species that rely Hickories (Carya) 235 Hemlocks (Tsuga) 92 Hollies (Ilex) 39 Poison Ivy (Toxicodendron) 15 locust, and Kentucky coffee trees. -

Anthelmintic Activity of Alangium Salviifolium Bark
Available online a t www.scholarsresearchlibrary.com Scholars Research Library J. Nat. Prod. Plant Resour ., 2012, 2 (6):717-720 (http://scholarsresearchlibrary.com/archive.html) ISSN : 2231 – 3184 CODEN (USA): JNPPB7 Anthelmintic activity of Alangium salviifolium bark Ravi Shankar Pandey SLT Institute of Pharmaceutical Sciences, Guru Ghasidas Vishwavidyalaya, Bilaspur, C.G _____________________________________________________________________________________________ ABSTRACT Alangium salviifolium (AS) is a novel medicinal plant used for the treatment of various diseases including helminthiasis by the traditional healers of Chhattisgarh. So an attempt has been taken to explore it scientifically. Preliminary phytochemical investigation reveals that there is presence of flavonoids, Saponins, phenols, bitter principles and steroids. Three different concentrations (50, 100 and 150 mg/ml) of crude extract of hexane, ethyl acetate, chloroform and methanol were tested against earthworms (Pheretima posthuma), which involved the evaluation of paralysis and death period of the worm. Methanol and chloroform extracts exhibited significant anthelmintic activity at highest concentration of 150 mg/mL. Piperzine citrate was selected as standard compound and DMF as control. The anthelmintic activity of Methanol and chloroform of bark of AS has therefore been evaluated for the first time. Key words: Alangium salviifolium; helminthiasis; Pheretima posthuma; Piperzine citrate and Paralysis _____________________________________________________________________________________________ INTRODUCTION Helminthiasis is a macroparasitic disease of humans and animals caused due to parasitic worms such as pinworm, roundworm, or tapeworm. These worms some times live in the gastrointestinal tract, may also burrow into the liver, lymphatic system, or other organs [1]. The clinical picture of consists of skin infection, skin nodules, and ocular lesions. The skin lesions include oedema, papules, scab-like eruptions and altered pigmentation. -

Likely to Have Habitat Within Iras That ALLOW Road
Item 3a - Sensitive Species National Master List By Region and Species Group Not likely to have habitat within IRAs Not likely to have Federal Likely to have habitat that DO NOT ALLOW habitat within IRAs Candidate within IRAs that DO Likely to have habitat road (re)construction that ALLOW road Forest Service Species Under NOT ALLOW road within IRAs that ALLOW but could be (re)construction but Species Scientific Name Common Name Species Group Region ESA (re)construction? road (re)construction? affected? could be affected? Bufo boreas boreas Boreal Western Toad Amphibian 1 No Yes Yes No No Plethodon vandykei idahoensis Coeur D'Alene Salamander Amphibian 1 No Yes Yes No No Rana pipiens Northern Leopard Frog Amphibian 1 No Yes Yes No No Accipiter gentilis Northern Goshawk Bird 1 No Yes Yes No No Ammodramus bairdii Baird's Sparrow Bird 1 No No Yes No No Anthus spragueii Sprague's Pipit Bird 1 No No Yes No No Centrocercus urophasianus Sage Grouse Bird 1 No Yes Yes No No Cygnus buccinator Trumpeter Swan Bird 1 No Yes Yes No No Falco peregrinus anatum American Peregrine Falcon Bird 1 No Yes Yes No No Gavia immer Common Loon Bird 1 No Yes Yes No No Histrionicus histrionicus Harlequin Duck Bird 1 No Yes Yes No No Lanius ludovicianus Loggerhead Shrike Bird 1 No Yes Yes No No Oreortyx pictus Mountain Quail Bird 1 No Yes Yes No No Otus flammeolus Flammulated Owl Bird 1 No Yes Yes No No Picoides albolarvatus White-Headed Woodpecker Bird 1 No Yes Yes No No Picoides arcticus Black-Backed Woodpecker Bird 1 No Yes Yes No No Speotyto cunicularia Burrowing -

The Vascular Plants of Massachusetts
The Vascular Plants of Massachusetts: The Vascular Plants of Massachusetts: A County Checklist • First Revision Melissa Dow Cullina, Bryan Connolly, Bruce Sorrie and Paul Somers Somers Bruce Sorrie and Paul Connolly, Bryan Cullina, Melissa Dow Revision • First A County Checklist Plants of Massachusetts: Vascular The A County Checklist First Revision Melissa Dow Cullina, Bryan Connolly, Bruce Sorrie and Paul Somers Massachusetts Natural Heritage & Endangered Species Program Massachusetts Division of Fisheries and Wildlife Natural Heritage & Endangered Species Program The Natural Heritage & Endangered Species Program (NHESP), part of the Massachusetts Division of Fisheries and Wildlife, is one of the programs forming the Natural Heritage network. NHESP is responsible for the conservation and protection of hundreds of species that are not hunted, fished, trapped, or commercially harvested in the state. The Program's highest priority is protecting the 176 species of vertebrate and invertebrate animals and 259 species of native plants that are officially listed as Endangered, Threatened or of Special Concern in Massachusetts. Endangered species conservation in Massachusetts depends on you! A major source of funding for the protection of rare and endangered species comes from voluntary donations on state income tax forms. Contributions go to the Natural Heritage & Endangered Species Fund, which provides a portion of the operating budget for the Natural Heritage & Endangered Species Program. NHESP protects rare species through biological inventory, -

2009 Wisconsin – Illinois Germplasm Reconnaissance and Collection Trips
NCRPIS 2009 Wisconsin – Illinois Germplasm Reconnaissance and Collection Trips August 3 – 8, 2009 September 21 – 29, 2009 North Central Regional Plant Introduction Station – Ames, IA Wisconsin Germplasm Sites Sauk County Washington County Cornus rugosa Fraxinus nigra (2) Fraxinus pennsylvanica Dane County Ilex verticillata Fraxinus americana Prunella vulgaris Fraxinus pennsylvanica Viburnum lentago Hypericum perforatum Spiraea alba Waukesha County Grant County Dasiphora fruticosa Cephalanthus occidentalis Fraxinus nigra Cornus alternifolia Fraxinus pennsylvanica Cornus rugosa Larix laricina Rock County Eupatorium maculatum Prunella vulgaris Fraxinus pennsylvanica Fraxinus pennsylvanica Dodge County Hypericum perforatum Cornus racemosa Monarda fistulosa Columbia County Fraxinus pennsylvanica Prunella vulgaris Aronia melanocarpa Prunella vulgaris Rudbeckia hirta Carpinus caroliniana Rudbeckia laciniata Cephalanthus occidentalis Jefferson County Staphylea trifolia Cornus amomum subsp. obliqua Fraxinus pennsylvanica Viburnum lentago Fraxinus nigra Fraxinus pennsylvanica Iowa County Ilex verticillata Diervilla lonicera Spiraea alba Fraxinus nigra Viburnum lentago Fraxinus pennsylvanica Rhus typhina Map of locations where germplasm was collected Illinois Germplasm Sites Winnebago County Ptelea trifoliata Cornus alternifolia Jo Daviess County Asclepias incarnata Clematis virginiana Cornus drummondii Fraxinus pennsylvanica Rudbeckia laciniata Spiraea alba Viburnum lentago Carroll County Rudbeckia laciniata Rudbeckia triloba Hypericum perforatum -

Outline of Angiosperm Phylogeny
Outline of angiosperm phylogeny: orders, families, and representative genera with emphasis on Oregon native plants Priscilla Spears December 2013 The following listing gives an introduction to the phylogenetic classification of the flowering plants that has emerged in recent decades, and which is based on nucleic acid sequences as well as morphological and developmental data. This listing emphasizes temperate families of the Northern Hemisphere and is meant as an overview with examples of Oregon native plants. It includes many exotic genera that are grown in Oregon as ornamentals plus other plants of interest worldwide. The genera that are Oregon natives are printed in a blue font. Genera that are exotics are shown in black, however genera in blue may also contain non-native species. Names separated by a slash are alternatives or else the nomenclature is in flux. When several genera have the same common name, the names are separated by commas. The order of the family names is from the linear listing of families in the APG III report. For further information, see the references on the last page. Basal Angiosperms (ANITA grade) Amborellales Amborellaceae, sole family, the earliest branch of flowering plants, a shrub native to New Caledonia – Amborella Nymphaeales Hydatellaceae – aquatics from Australasia, previously classified as a grass Cabombaceae (water shield – Brasenia, fanwort – Cabomba) Nymphaeaceae (water lilies – Nymphaea; pond lilies – Nuphar) Austrobaileyales Schisandraceae (wild sarsaparilla, star vine – Schisandra; Japanese -
![Alangium-Presentation [Lecture Seule]](https://docslib.b-cdn.net/cover/3512/alangium-presentation-lecture-seule-243512.webp)
Alangium-Presentation [Lecture Seule]
A Targeted Enrichment Strategy for Sequencing of Medicinal Species in the Indonesian Flora Berenice Villegas-Ramirez, Erasmus Mundus Master Programme in Evolutionary Biology (MEME) Supervisors: Dr. Sarah Mathews, Harvard University Dr. Hugo de Boer, Uppsala University Introduction • Up to 70,000 plant species are used worldwide in traditional medicine. • At least 20,000 plant taxa have recorded medicinal uses. • Main commercial producers are in Asia: China, India, Indonesia, and Nepal. • Indonesia has c. 7000 plant species of documented medicinal use. • But…… Transmigration and Farming Herbarium Specimens • Plastid genes rbcL and matK have been be adopted as the official DNA barcodes for all land plants. • rbcl ~ 1428 bp • matK ~ 1500 bp • Herbarium specimens often require more attempts at amplification with more primer combinations. • Higher possibility of obtaining incorrect sequences through increased chances of samples becoming mixed up or contaminated. • Lower performance using herbarium material due to lower amplification success. • Caused by severe degradation of DNA into low molecular weight fragments. • But fragmented DNA is not a curse! Next-Generation Sequencing • Fragmented DNA is less of a problem • Only a few milligrams of material are necessary Targeted Enrichment • Defined regions in a genome are selectively captured from a DNA sample prior to sequencing. • The genomic complexity in a sample is reduced. • More time- and cost-effective. Hybrid Capture Targeted Enrichment • Library DNA is hybridized to a probe. • Pre-prepared DNA or RNA fragments complementary to the targeted regions of interest. • Non-specific hybrids are removed by washing. • Targeted DNA is eluted. Easy to use, utilizes a small amount of input DNA (<1-3 ug), and number of loci (target size) is large (1-50 Mb). -

Tagawa Gardens June Snow Giant Dogwood
June Snow Giant Dogwood Cornus controversa 'June Snow-JFS' Height: 30 feet Spread: 40 feet Sunlight: Hardiness Zone: 4 Description: June Snow Giant Dogwood flowers Covered with lovely white flower clusters in spring; a Photo courtesy of NetPS Plant Finder large rounded form, with gracefully layered branches make this tree an excellent specimen; attractive fruit in late summer turns deep blue-black; orange to red fall color Ornamental Features June Snow Giant Dogwood features showy clusters of white flowers held atop the branches in late spring. It has dark green foliage throughout the season. The pointy leaves turn an outstanding dark red in the fall. It produces navy blue berries from early to late fall. The warty gray bark and green branches add an interesting dimension to the landscape. June Snow Giant Dogwood in bloom Landscape Attributes Photo courtesy of NetPS Plant Finder June Snow Giant Dogwood is a deciduous tree with a stunning habit of growth which features almost oriental horizontally-tiered branches. Its average texture blends into the landscape, but can be balanced by one or two finer or coarser trees or shrubs for an effective composition. This is a relatively low maintenance tree, and should only be pruned after flowering to avoid removing any of the current season's flowers. It is a good choice for attracting birds to your yard. It has no significant negative characteristics. June Snow Giant Dogwood is recommended for the following landscape applications; - Accent - Shade Planting & Growing June Snow Giant Dogwood will grow to be about 30 feet tall at maturity, with a spread of 40 feet. -

A Stochastic Model of Language Evolution That Incorporates Homoplasy and Borrowing
A STOCHASTIC MODEL OF LANGUAGE EVOLUTION THAT INCORPORATES HOMOPLASY AND BORROWING TANDY WARNOW, STEVEN N. EVANS, DONALD RINGE, AND LUAY NAKHLEH 1. Introduction The inference of evolutionary history, whether in biology or in lin- guistics, is aided by a carefully considered model of the evolutionary process and a reconstruction method which is expected to produce a reasonably accurate estimation of the true evolutionary history when the real data match the model assumptions and are of sufficient quan- tity. In molecular systematics (i.e., the inference of evolutionary his- tories from molecular data), much of the research effort has focused in two areas: first, the development of increasingly parameter rich models of molecular sequence evolution, and second, the development of increasingly sophisticated software tools and algorithms for recon- structing phylogenies under these models. The plethora of software for reconstructing phylogenies from molecular data is staggering. By com- parison, much less has been done in historical linguistics in terms of developing statistical models of character evolution or reconstruction methods, suggesting that there is perhaps much to be gained by doing so. To date, although some models have been proposed for language evo- lution, all have failed in some significant ways. In particular, linguistic models either explicitly or implicitly have assumed that no homoplasy (i.e., parallel evolution and/or back–mutation) occurs (see for exam- ple (Ringe et al. , 2002; Taylor et al. , 2000; Warnow, 1997)). Most, but not all, have not modelled borrowing between languages. In this paper, we go beyond earlier models by explicitly incorporating both homoplasy and borrowing into our model.