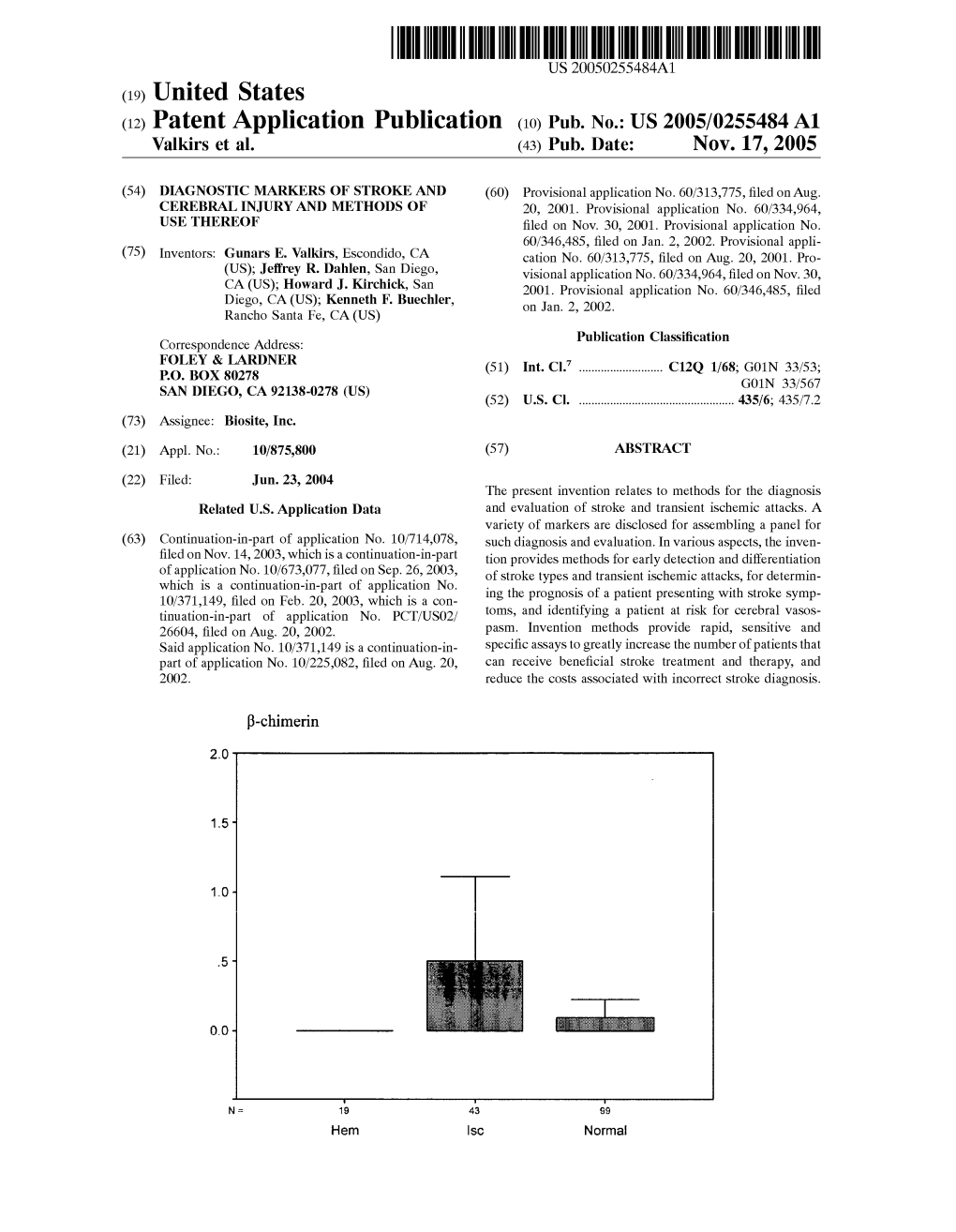
(12) Patent Application Publication (10) Pub. No.: US 2005/0255484A1 Walkirs Et Al
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Identification of a Novel CHN1 P.(Phe213val) Variant in a Large Han Chinese Family with Congenital Duane Retraction Syndrome
www.nature.com/scientificreports OPEN Identifcation of a novel CHN1 p.(Phe213Val) variant in a large Han Chinese family with congenital Duane retraction syndrome Tai‑Cheng Zhou1,3, Wen‑Hua Duan1,3, Xiao‑Lin Fu2,3, Qin Zhu1, Li‑Yun Guo1, Yuan Zhou1, Zhi‑Juan Hua1, Xue‑Jiao Li1, Dong‑Mei Yang1, Jie‑Ying Zhang1, Jie Yin1, Xiao‑Fan Zhang1, Guang‑Long Zhou1 & Min Hu1* Duane retraction syndrome (DRS) is a neuromuscular dysfunction of the eyes. Although many causative genes of DRS have been identifed in Europe and the United States, few reports have been published in regard to Chinese DRS. The aim of the present study was to explore the genetic defect of DRS in a Chinese family. Exome sequencing was used to identify the disease‑causing gene for the two afected family members. Ophthalmic and physical examinations, as well as genetic screenings for variants in chimerin 1 (CHN1), were performed for all family members. Functional analyses of a CHN1 variant in 293T cells included a Rac‑GTP activation assay, α2‑chimaerin translocation assay, and co‑immunoprecipitation assay. Genetic analysis revealed a NM_001822.7: c.637T > G variant in the CHN1 gene, which resulted in the substitution of a highly conserved C1 domain with valine at codon 213 (NP_001813.1: p.(Phe213Val)) (ClinVar Accession Number: SCV001335305). In-silico analysis revealed that the p.(Phe213Val) substitution afected the protein stability and connections among the amino acids of CHN1 in terms of its tertiary protein structure. Functional studies indicated that the p.(Phe213Val) substitution reduced Rac‑GTP activity and enhanced membrane translocation in response to phorbol‑myristoyl acetate (PMA). -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Cellular and Molecular Signatures in the Disease Tissue of Early
Cellular and Molecular Signatures in the Disease Tissue of Early Rheumatoid Arthritis Stratify Clinical Response to csDMARD-Therapy and Predict Radiographic Progression Frances Humby1,* Myles Lewis1,* Nandhini Ramamoorthi2, Jason Hackney3, Michael Barnes1, Michele Bombardieri1, Francesca Setiadi2, Stephen Kelly1, Fabiola Bene1, Maria di Cicco1, Sudeh Riahi1, Vidalba Rocher-Ros1, Nora Ng1, Ilias Lazorou1, Rebecca E. Hands1, Desiree van der Heijde4, Robert Landewé5, Annette van der Helm-van Mil4, Alberto Cauli6, Iain B. McInnes7, Christopher D. Buckley8, Ernest Choy9, Peter Taylor10, Michael J. Townsend2 & Costantino Pitzalis1 1Centre for Experimental Medicine and Rheumatology, William Harvey Research Institute, Barts and The London School of Medicine and Dentistry, Queen Mary University of London, Charterhouse Square, London EC1M 6BQ, UK. Departments of 2Biomarker Discovery OMNI, 3Bioinformatics and Computational Biology, Genentech Research and Early Development, South San Francisco, California 94080 USA 4Department of Rheumatology, Leiden University Medical Center, The Netherlands 5Department of Clinical Immunology & Rheumatology, Amsterdam Rheumatology & Immunology Center, Amsterdam, The Netherlands 6Rheumatology Unit, Department of Medical Sciences, Policlinico of the University of Cagliari, Cagliari, Italy 7Institute of Infection, Immunity and Inflammation, University of Glasgow, Glasgow G12 8TA, UK 8Rheumatology Research Group, Institute of Inflammation and Ageing (IIA), University of Birmingham, Birmingham B15 2WB, UK 9Institute of -

Ck1δ Over-Expressing Mice Display ADHD-Like Behaviors, Frontostriatal Neuronal Abnormalities and Altered Expressions of ADHD-Candidate Genes
Molecular Psychiatry (2020) 25:3322–3336 https://doi.org/10.1038/s41380-018-0233-z ARTICLE CK1δ over-expressing mice display ADHD-like behaviors, frontostriatal neuronal abnormalities and altered expressions of ADHD-candidate genes 1 1 1 2 1 1 1 Mingming Zhou ● Jodi Gresack ● Jia Cheng ● Kunihiro Uryu ● Lars Brichta ● Paul Greengard ● Marc Flajolet Received: 8 November 2017 / Revised: 4 July 2018 / Accepted: 18 July 2018 / Published online: 19 October 2018 © Springer Nature Limited 2018 Abstract The cognitive mechanisms underlying attention-deficit hyperactivity disorder (ADHD), a highly heritable disorder with an array of candidate genes and unclear genetic architecture, remain poorly understood. We previously demonstrated that mice overexpressing CK1δ (CK1δ OE) in the forebrain show hyperactivity and ADHD-like pharmacological responses to D- amphetamine. Here, we demonstrate that CK1δ OE mice exhibit impaired visual attention and a lack of D-amphetamine- induced place preference, indicating a disruption of the dopamine-dependent reward pathway. We also demonstrate the presence of abnormalities in the frontostriatal circuitry, differences in synaptic ultra-structures by electron microscopy, as 1234567890();,: 1234567890();,: well as electrophysiological perturbations of both glutamatergic and GABAergic transmission, as observed by altered frequency and amplitude of mEPSCs and mIPSCs. Furthermore, gene expression profiling by next-generation sequencing alone, or in combination with bacTRAP technology to study specifically Drd1a versus Drd2 medium spiny neurons, revealed that developmental CK1δ OE alters transcriptional homeostasis in the striatum, including specific alterations in Drd1a versus Drd2 neurons. These results led us to perform a fine molecular characterization of targeted gene networks and pathway analysis. Importantly, a large fraction of 92 genes identified by GWAS studies as associated with ADHD in humans are significantly altered in our mouse model. -

Neuronal and Glial DNA Methylation and Gene Expression Changes in Early Epileptogenesis
RESEARCH ARTICLE Neuronal and glial DNA methylation and gene expression changes in early epileptogenesis 1,2☯ 3☯ 3 4,5 Toni C. BergerID *, Magnus D. Vigeland , Hanne S. Hjorthaug , Lars Etholm , Cecilie G. Nome2, Erik Taubøll1,2, Kjell Heuser1,2³, Kaja K. Selmer3,4,6³ 1 Department of Neurology, Oslo University Hospital, Oslo, Norway, 2 University of Oslo, Oslo, Norway, 3 Department of Medical Genetics, Oslo University Hospital and University of Oslo, Oslo, Norway, 4 National Center for Epilepsy, Oslo University Hospital, Sandvika, Norway, 5 Department of Neurology, Section for Neurophysiology, Oslo University Hospital, Oslo, Norway, 6 Division of Clinical Neuroscience, Department of Research and Development, Oslo University Hospital, Oslo, Norway a1111111111 ☯ These authors contributed equally to this work. a1111111111 ³ KH and KKS also contributed equally to this work. a1111111111 * [email protected] a1111111111 a1111111111 Abstract Background and aims OPEN ACCESS Mesial Temporal Lobe Epilepsy is characterized by progressive changes of both neurons Citation: Berger TC, Vigeland MD, Hjorthaug HS, Etholm L, Nome CG, Taubøll E, et al. (2019) and glia, also referred to as epileptogenesis. No curative treatment options, apart from sur- Neuronal and glial DNA methylation and gene gery, are available. DNA methylation (DNAm) is a potential upstream mechanism in epilep- expression changes in early epileptogenesis. PLoS togenesis and may serve as a novel therapeutic target. To our knowledge, this is the first ONE 14(12): e0226575. https://doi.org/10.1371/ study to investigate epilepsy-related DNAm, gene expression (GE) and their relationship, in journal.pone.0226575 neurons and glia. Editor: Giuseppe Biagini, University of Modena and Reggio Emilia, ITALY Methods Received: July 18, 2019 We used the intracortical kainic acid injection model to elicit status epilepticus. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

Identification of Potential Key Genes and Pathway Linked with Sporadic Creutzfeldt-Jakob Disease Based on Integrated Bioinformatics Analyses
medRxiv preprint doi: https://doi.org/10.1101/2020.12.21.20248688; this version posted December 24, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in perpetuity. All rights reserved. No reuse allowed without permission. Identification of potential key genes and pathway linked with sporadic Creutzfeldt-Jakob disease based on integrated bioinformatics analyses Basavaraj Vastrad1, Chanabasayya Vastrad*2 , Iranna Kotturshetti 1. Department of Biochemistry, Basaveshwar College of Pharmacy, Gadag, Karnataka 582103, India. 2. Biostatistics and Bioinformatics, Chanabasava Nilaya, Bharthinagar, Dharwad 580001, Karanataka, India. 3. Department of Ayurveda, Rajiv Gandhi Education Society`s Ayurvedic Medical College, Ron, Karnataka 562209, India. * Chanabasayya Vastrad [email protected] Ph: +919480073398 Chanabasava Nilaya, Bharthinagar, Dharwad 580001 , Karanataka, India NOTE: This preprint reports new research that has not been certified by peer review and should not be used to guide clinical practice. medRxiv preprint doi: https://doi.org/10.1101/2020.12.21.20248688; this version posted December 24, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in perpetuity. All rights reserved. No reuse allowed without permission. Abstract Sporadic Creutzfeldt-Jakob disease (sCJD) is neurodegenerative disease also called prion disease linked with poor prognosis. The aim of the current study was to illuminate the underlying molecular mechanisms of sCJD. The mRNA microarray dataset GSE124571 was downloaded from the Gene Expression Omnibus database. Differentially expressed genes (DEGs) were screened. -

Signal Transduction in L-DOPA-Induced Dyskinesia: from Receptor Sensitization to Abnormal… Et Al
Journal of Neural Transmission https://doi.org/10.1007/s00702-018-1847-7 NEUROLOGY AND PRECLINICAL NEUROLOGICAL STUDIES - REVIEW ARTICLE Signal transduction in L‑DOPA‑induced dyskinesia: from receptor sensitization to abnormal gene expression Giada Spigolon1 · Gilberto Fisone1 Received: 1 November 2017 / Accepted: 23 January 2018 © The Author(s) 2018. This article is an open access publication Abstract A large number of signaling abnormalities have been implicated in the emergence and expression of L-DOPA-induced dyskinesia (LID). The primary cause for many of these changes is the development of sensitization at dopamine receptors located on striatal projection neurons (SPN). This initial priming, which is particularly evident at the level of dopamine D1 receptors (D1R), can be viewed as a homeostatic response to dopamine depletion and is further exacerbated by chronic administration of L-DOPA, through a variety of mechanisms afecting various components of the G-protein-coupled receptor machinery. Sensitization of dopamine receptors in combination with pulsatile administration of L-DOPA leads to intermit- tent and coordinated hyperactivation of signal transduction cascades, ultimately resulting in long-term modifcations of gene expression and protein synthesis. A detailed mapping of these pathological changes and of their involvement in LID has been produced during the last decade. According to this emerging picture, activation of sensitized D1R results in the stimulation of cAMP-dependent protein kinase and of the dopamine- and cAMP-regulated phosphoprotein of 32 kDa. This, in turn, activates the extracellular signal-regulated kinases 1 and 2 (ERK), leading to chromatin remodeling and aberrant gene transcription. Dysregulated ERK results also in the stimulation of the mammalian target of rapamycin complex 1, which promotes protein synthesis. -

Systematic Analysis of Brain Lactate and Ph Levels in 65 Animal Models Related To
bioRxiv preprint doi: https://doi.org/10.1101/2021.02.02.428362; this version posted February 4, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Full title: Systematic analysis of brain lactate and pH levels in 65 animal models related to 2 neuropsychiatric conditions 3 Short title: Brain lactate and pH in neuropsychiatric disorder models 4 Authors: Hideo Hagihara1,2, Hirotaka Shoji1,2, International Brain pH Project Consortium2, 5 and Tsuyoshi Miyakawa1,2* 6 Affiliations: 7 1 Division of Systems Medical Science, Institute for Comprehensive Medical Science, 8 Fujita Health University, Toyoake, Aichi 470-1192, Japan 9 2A full list of the authors in the International Brain pH Project Consortium is provided 10 at the end of the paper along with their affiliations and contributions. 11 *Corresponding Author: Tsuyoshi Miyakawa ([email protected]) 12 Keywords: lactate, pH, brain, neuropsychiatric disorders, animal models, working memory 13 bioRxiv preprint doi: https://doi.org/10.1101/2021.02.02.428362; this version posted February 4, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 14 Abstract 15 Altered brain energy metabolism associated with increase in lactate levels and the 16 resultant decrease in pH have been increasingly implicated in multiple neuropsychiatric 17 disorders, such as schizophrenia, bipolar disorder, autism spectrum disorder and 18 neurodegenerative disorders. -
Supplemental Data Olig2-Regulated Lineage-Restricted Pathway Controls Replication Competence in Neural Stem Cells and Malignant
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Caltech Authors Neuron, Volume 53 Supplemental Data Olig2-Regulated Lineage-Restricted Pathway Controls Replication Competence in Neural Stem Cells and Malignant Glioma Keith L. Ligon, Emmanuelle Huillard, Shwetal Mehta, Santosh Kesari, Hongye Liu, John A. Alberta, Robert M. Bachoo, Michael Kane, David N. Louis, Ronald A. DePinho, David J. Anderson, Charles D. Stiles, and David H. Rowitch Figure S1. Olig1/2+/- Ink4a/Arf-/-EGFRvIII Gliomas Express Characteristic Morphologic and Immunophenotypic Features of Human Malignant Gliomas (A) Tumors exhibit dense cellularity and atypia. (B) Characteristic infiltration of host SVZ stem cell niche. (C) Although occasional tumors exhibited pseudopalisading necrosis (n, arrowhead) and hemorrhage (h, arrowhead) similar to human GBM (Astrocytoma WHO Grade IV), most tumors lacked these features. (D) IHC for hEGFR highlights hallmark feature of human gliomas, including perivascular (pv) and subpial (sp) accumulation of tumor cells, (E) striking white matter tropism of tumor cells in corpus callosum (cc), and (F) distant single cell infiltration of cerebellar white matter (wm, arrows). (G-J) Immunohistochemical markers characteristic of human tumors (Gfap, Olig2, Olig1, Nestin) are present. (K and L) Weak staining for the early neuronal marker TuJ1 and absence of the differentiated neuronal marker, NeuN similar to human glioma and consistent with heterogeneous lines of partial differentiation. Figure S2. Olig1/2+/- Ink4a/Arf-/-EGFRvIII Neurospheres Are Multipotent and Olig1/2-/- Ink4a/Arf-/-EGFRvIII Neurospheres Are Bipotent Olig1/2+/- or Olig1/2-/- Ink4a/Arf-/-EGFRvIII neurospheres were allowed to differentiate for 6 days in medium without EGF. -

Supplementary Materials
Supplementary material. S1. Images from automatic imaging reader Cytation™ 1. A, A’, A’’ present the same field of view. Cells migrating from the upper compartment of the inserts are stained with DID (red color), Cell nuclei are stained with DAPI (blue color). A’ and A’’ demonstrate the method of analysis (A’ – nuclei numbering, A’’ – migrating cells numbering), scale bar – 300 µm. ASC BM-MSC Y BM-MSC A mean SEM mean SEM mean SEM CXCL6 2.240 0.727 4.158 0.677 2.149 1.816 CXCL16 4.277 0.248 0.763 0.405 1.130 0.346 CXCL12 -2.855 0.483 -4.528 0.226 -3.616 0.318 SMAD3 -0.511 0.191 -1.522 0.188 -1.332 0.215 TGFB2 3.742 0.533 1.238 0.210 0.990 0.568 COL14A 1.532 0.357 -0.397 0.736 -1.522 0.469 MHX 1.397 0.414 1.145 0.412 0.642 0.613 SCX 2.984 0.301 2.062 0.320 2.031 0.249 RUNX2 3.290 0.359 2.319 0.388 2.546 0.529 PPRAG 1.720 0.303 2.926 0.423 2.912 0.215 Supplementary material S3. The table provides the mean Δct and SEM values for all genes and all groups analyzed in this study (n=8 in each group). Supplementary material S2. The table provides the list of genes which displayed significantly different expression between hASCs and hBM-MSCs A based on microarray nr Exp p-value Exp Fold Change ID Symbol Entrez Gene Name 1 2.84E-05 16.896 16960355 TM4SF1 transmembrane 4 L six family member 1 2 9.10E-09 12.238 16684080 IFI6 interferon alpha inducible protein 6 3 3.93E-08 11.851 16667702 VCAM1 vascular cell adhesion molecule 1 4 1.54E-05 9.944 16840113 CXCL16 C-X-C motif chemokine ligand 16 5 2.01E-06 9.123 16852463 RAB27B RAB27B, member RAS oncogene -

Supplementary Table 2
Supplementary Table 2. Differentially Expressed Genes following Sham treatment relative to Untreated Controls Fold Change Accession Name Symbol 3 h 12 h NM_013121 CD28 antigen Cd28 12.82 BG665360 FMS-like tyrosine kinase 1 Flt1 9.63 NM_012701 Adrenergic receptor, beta 1 Adrb1 8.24 0.46 U20796 Nuclear receptor subfamily 1, group D, member 2 Nr1d2 7.22 NM_017116 Calpain 2 Capn2 6.41 BE097282 Guanine nucleotide binding protein, alpha 12 Gna12 6.21 NM_053328 Basic helix-loop-helix domain containing, class B2 Bhlhb2 5.79 NM_053831 Guanylate cyclase 2f Gucy2f 5.71 AW251703 Tumor necrosis factor receptor superfamily, member 12a Tnfrsf12a 5.57 NM_021691 Twist homolog 2 (Drosophila) Twist2 5.42 NM_133550 Fc receptor, IgE, low affinity II, alpha polypeptide Fcer2a 4.93 NM_031120 Signal sequence receptor, gamma Ssr3 4.84 NM_053544 Secreted frizzled-related protein 4 Sfrp4 4.73 NM_053910 Pleckstrin homology, Sec7 and coiled/coil domains 1 Pscd1 4.69 BE113233 Suppressor of cytokine signaling 2 Socs2 4.68 NM_053949 Potassium voltage-gated channel, subfamily H (eag- Kcnh2 4.60 related), member 2 NM_017305 Glutamate cysteine ligase, modifier subunit Gclm 4.59 NM_017309 Protein phospatase 3, regulatory subunit B, alpha Ppp3r1 4.54 isoform,type 1 NM_012765 5-hydroxytryptamine (serotonin) receptor 2C Htr2c 4.46 NM_017218 V-erb-b2 erythroblastic leukemia viral oncogene homolog Erbb3 4.42 3 (avian) AW918369 Zinc finger protein 191 Zfp191 4.38 NM_031034 Guanine nucleotide binding protein, alpha 12 Gna12 4.38 NM_017020 Interleukin 6 receptor Il6r 4.37 AJ002942