Outlier Detection in the Statistical Accuracy Test of a Pka Prediction
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Location-Scale Distributions
Location–Scale Distributions Linear Estimation and Probability Plotting Using MATLAB Horst Rinne Copyright: Prof. em. Dr. Horst Rinne Department of Economics and Management Science Justus–Liebig–University, Giessen, Germany Contents Preface VII List of Figures IX List of Tables XII 1 The family of location–scale distributions 1 1.1 Properties of location–scale distributions . 1 1.2 Genuine location–scale distributions — A short listing . 5 1.3 Distributions transformable to location–scale type . 11 2 Order statistics 18 2.1 Distributional concepts . 18 2.2 Moments of order statistics . 21 2.2.1 Definitions and basic formulas . 21 2.2.2 Identities, recurrence relations and approximations . 26 2.3 Functions of order statistics . 32 3 Statistical graphics 36 3.1 Some historical remarks . 36 3.2 The role of graphical methods in statistics . 38 3.2.1 Graphical versus numerical techniques . 38 3.2.2 Manipulation with graphs and graphical perception . 39 3.2.3 Graphical displays in statistics . 41 3.3 Distribution assessment by graphs . 43 3.3.1 PP–plots and QQ–plots . 43 3.3.2 Probability paper and plotting positions . 47 3.3.3 Hazard plot . 54 3.3.4 TTT–plot . 56 4 Linear estimation — Theory and methods 59 4.1 Types of sampling data . 59 IV Contents 4.2 Estimators based on moments of order statistics . 63 4.2.1 GLS estimators . 64 4.2.1.1 GLS for a general location–scale distribution . 65 4.2.1.2 GLS for a symmetric location–scale distribution . 71 4.2.1.3 GLS and censored samples . -

An Introduction to R Notes on R: a Programming Environment for Data Analysis and Graphics Version 4.1.1 (2021-08-10)
An Introduction to R Notes on R: A Programming Environment for Data Analysis and Graphics Version 4.1.1 (2021-08-10) W. N. Venables, D. M. Smith and the R Core Team This manual is for R, version 4.1.1 (2021-08-10). Copyright c 1990 W. N. Venables Copyright c 1992 W. N. Venables & D. M. Smith Copyright c 1997 R. Gentleman & R. Ihaka Copyright c 1997, 1998 M. Maechler Copyright c 1999{2021 R Core Team Permission is granted to make and distribute verbatim copies of this manual provided the copyright notice and this permission notice are preserved on all copies. Permission is granted to copy and distribute modified versions of this manual under the conditions for verbatim copying, provided that the entire resulting derived work is distributed under the terms of a permission notice identical to this one. Permission is granted to copy and distribute translations of this manual into an- other language, under the above conditions for modified versions, except that this permission notice may be stated in a translation approved by the R Core Team. i Table of Contents Preface :::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::: 1 1 Introduction and preliminaries :::::::::::::::::::::::::::::::: 2 1.1 The R environment :::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::: 2 1.2 Related software and documentation ::::::::::::::::::::::::::::::::::::::::::::::: 2 1.3 R and statistics :::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::: 2 1.4 R and the window system :::::::::::::::::::::::::::::::::::::::::::::::::::::::::: -

SEQUENTIAL PROCEDURES in PROBIT ANALYSIS by TADEPALLI
SEQUENTIAL PROCEDURES IN PROBIT ANALYSIS by TADEPALLI VENKATA NARAYANA Special report to THE UNITED STATES AIR FORCE under Contract AF 18(600)-83 monitored by the Office of Scientific Research. Institute of Statistics Mimeograph Series No.82 October, 1953 SEQUENTIAL PROCEDURES IN PROBIT ANALYSIS by TADEPALLI VENKATA NARAYANA A thesis submitted to the Faculty of the University of North Carolina in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Department of Statistics Chapel Hill 1953 Approved by: Adviser ii ACK NOW LED GEM ENT I wish to express my deep gratitude to Professor N. L. Johnson for suggesting the problem and constantly guiding me throughout the preparation of this dissertation. I wish also to thank the Institute of Statistics and the U. S. Air Force for financial assistance which made this study possible. T. V. Narayana iii TABLE OF CONTENTS Page ACKNOWLEDGEMENT ii INTRODUCTION iv Chapter I. STATm1~NT OF PROBLEM 1 II. APPLICATION OF THE MOOD AND DIXON HETHOD TO RANKITS 14 III. THE ALTERNATIVE METHODS AND THE MOOD AND DDCON METHOD IN THE CASE OF SMALL SAMPLES 23 IV. THE ASYMPTOTIC PROPERTIES OF THE DIFFERENT SEnUENTIAL PROCEDURES 48 v. ESTIMATION OF THE ED,O IN SAMPLES OF MODERATE SIZES 79 APPENDIX 97 BIBLIOGRAPHY 107 INTRODUCTION The standard technique of Probit Analysis enables us to es timate the "median effective dose" (ED50) in a biological assay. When a simple normalizing transformation for the doses is avail able, the normalizing measure of "dosage" has a normally distribu ted tolerance. The problem of estimating the mean and standard deviation of this distribution is solved by the probit transforma tion of the experimental results. -

Statistix R at Clemson University
R Statistix at Clemson University Marie Con Herman Senter Contents 1 Intro duction { Read Me First! 1 1.1 Starting the program : : : : : : : : : : : : : : : : : : : : : : : 1 1.2 Selecting a dataset : : : : : : : : : : : : : : : : : : : : : : : : 2 1.3 Finding your way around : : : : : : : : : : : : : : : : : : : : : 2 1.4 Changing your mind : : : : : : : : : : : : : : : : : : : : : : : 3 1.5 Printing : : : : : : : : : : : : : : : : : : : : : : : : : : : : : : 3 1.6 Exiting the Program : : : : : : : : : : : : : : : : : : : : : : : 3 1.7 Note : : : : : : : : : : : : : : : : : : : : : : : : : : : : : : : : 4 2 Graphs 5 2.1 Histograms and Bar Charts : : : : : : : : : : : : : : : : : : : 5 2.1.1 Mo di cations : : : : : : : : : : : : : : : : : : : : : : : 6 2.1.2 A normal curve : : : : : : : : : : : : : : : : : : : : : : 7 2.2 Stem-and-leaf Diagrams (Stemplots) : : : : : : : : : : : : : : 7 2.3 Box-and-Whisker Plot (Boxplot) : : : : : : : : : : : : : : : : : 8 2.3.1 Side-by-side b oxplots : : : : : : : : : : : : : : : : : : : 8 2.4 Error Bar Charts : : : : : : : : : : : : : : : : : : : : : : : : : 10 2.5 Scatterplots (X-Y plots) : : : : : : : : : : : : : : : : : : : : : 12 2.5.1 Plotting a line : : : : : : : : : : : : : : : : : : : : : : : 13 2.6 Control Charts : : : : : : : : : : : : : : : : : : : : : : : : : : 13 2.7 Time plots : : : : : : : : : : : : : : : : : : : : : : : : : : : : : 13 2.8 Normal probability plots (Normal quantile plots) : : : : : : : 13 3 Descriptive Statistics 15 3.1 Grouping variables : : : : : : : : : : : : : : : : -

5 Checking Assumptions
5 CHECKING ASSUMPTIONS 5 Checking Assumptions Almost all statistical methods make assumptions about the data collection process and the shape of the population distribution. If you reject the null hypothesis in a test, then a reasonable conclusion is that the null hypothesis is false, provided all the distributional assumptions made by the test are satisfied. If the assumptions are not satisfied then that alone might be the cause of rejecting H0. Additionally, if you fail to reject H0, that could be caused solely by failure to satisfy assumptions also. Hence, you should always check assumptions to the best of your abilities. Two assumptions that underly the tests and CI procedures that I have discussed are that the data are a random sample, and that the population frequency curve is normal. For the pooled variance two sample test the population variances are also required to be equal. The random sample assumption can often be assessed from an understanding of the data col- lection process. Unfortunately, there are few general tests for checking this assumption. I have described exploratory (mostly visual) methods to assess the normality and equal variance assump- tion. I will now discuss formal methods to assess these assumptions. Testing Normality A formal test of normality can be based on a normal scores plot, sometimes called a rankit plot or a normal probability plot or a normal Q-Q plot. You plot the data against the normal scores, or expected normal order statistics (in a standard normal) for a sample with the given number of observations. The normality assumption is plausible if the plot is fairly linear. -
Testing Assumptions: Normality and Equal Variances
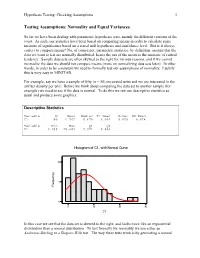
Hypothesis Testing: Checking Assumptions 1 Testing Assumptions: Normality and Equal Variances So far we have been dealing with parametric hypothesis tests, mainly the different versions of the t-test. As such, our statistics have been based on comparing means in order to calculate some measure of significance based on a stated null hypothesis and confidence level. But is it always correct to compare means? No, of course not; parametric statistics, by definition, assume that the data we want to test are normally distributed, hence the use of the mean as the measure of central tendency. Sample data sets are often skewed to the right for various reasons, and if we cannot normalize the data we should not compare means (more on normalizing data sets later). In other words, in order to be consistent we need to formally test our assumptions of normality. Luckily this is very easy in MINITAB. For example, say we have a sample of fifty (n = 50) excavated units and we are interested in the artifact density per unit. Before we think about comparing the data set to another sample (for example) we need to see if the data is normal. To do this we run our descriptive statistics as usual and produce some graphics: Descriptive Statistics Variable N Mean Median Tr Mean StDev SE Mean C1 50 6.702 5.679 6.099 5.825 0.824 Variable Min Max Q1 Q3 C1 0.039 25.681 2.374 9.886 Histogram of C1, with Normal Curve 10 ncy que 5 Fre 0 0 10 20 30 C1 In this case we see that the data set is skewed to the right, and looks more like an exponential distribution than a normal distribution. -

Lecture Notes (/Wws509/Notes)
2015/9/12 WWS 509 Germán Rodríguez Generalized Linear Models Lecture Notes (/wws509/notes) Lecture Notes The notes are offered in two formats: HTML and PDF. I expect most of you will want to print the notes, in which case you can use the links below to access the PDF file for each chapter. If you are browsing use the table of contents to jump directly to each chapter and section in HTML format. For more details on these formats please see the discussion below. Chapters in PDF Format 2. Linear Models for Continuous Data (c2.pdf) 3. Logit Models for Binary Data (c3.pdf) 4. Poisson Models for Count Data (c4.pdf) 4a*. Addendun on Overdispersed Count Data (c4a.pdf) 5. Log-Linear Models for Contingency Tables (c5.pdf) 6. Multinomial Response Models (c6.pdf) 7. Survival Models (c7.pdf) 8*. Panel and Clustered Data (fixedRandom.pdf) A. Review of Likelihood Theory (a1.pdf) B. Generalized Linear Model Theory (a2.pdf) The list above has two extensions to the original notes: an addendum (c4addendum.pdf) on Over- Dispersed Count Data, which describes models with extra-Poisson variation and negative binomial regression, and a brief discussion (fixedRandom.pdf) of models for longitudinal and clustered data. Because of these additions we now skip Chapter 5. http://data.princeton.edu/wws509/notes 1/2 2015/9/12 WWS 509 No, there is no Chapter 1 ... yet. One day I will write an introduction to the course and that will be Chapter 1. The Choice of Formats It turns out that making the lecture notes available on the web was a bit of a challenge because web browsers were designed to render text and graphs but not equations, which are often shown using bulky graphs or translated into text with less than ideal results. -

Glosario EN-ES De Ensayos Clínicos (2.ª Parte: N-Z)
<http://tremedica.org/panacea.html> Traducción y terminología Glosario EN-ES de ensayos clínicos (2.ª parte: N-Z). M.ª Verónica Saladrigas,* Fernando A. Navarro,** Laura Munoa,*** Pablo Mugüerza**** y Álvaro Villegas*****. Resumen: La investigación clínica, y dentro de ella la basada en ensayos clínicos con medicamentos, genera un volumen ingente de documentación escrita que es preciso traducir del inglés al español. El presente glosario está pensado como ayuda práctica al traductor especializado que se enfrenta a esta compleja tarea. Se han seleccionado cerca de 1400 conceptos básicos del ámbito de los ensayos clínicos y otras disciplinas afines y se han organizado buscando la máxima claridad expositiva. Para cada entrada principal se ofrecen uno o más equivalentes en español, seleccionados según criterios que tienen en cuenta tanto la frecuencia de uso real como la corrección lingüística y conceptual. El glosario se enriquece con unas 1500 remisiones internas a voces equivalentes o a entradas relacionadas. Por último, muchos artículos aportan información complementaria de interés para el traductor. La presente entrega contiene las entradas de la N a la Z. Palabras clave: ensayo clínico; español; inglés; investigación clínica. Glossary of clinical trials, ENG-SPA (2nd part: N-Z). Abstract: Clinical research, including the investigation based on pharmacological clinical trials, produces an enormous amount of written documentation that has to be translated from English into Spanish. This glossary is intended as a practical guide for the specialized translator who faces this complex task. About 1,400 basic concepts in the clinical trial environment and other related disciplines have been selected and organ- ized with the purpose of achieving an optimal content clarity. -

Using R at the Bench: Step-By-Step Analytics for Biologists
This is a free sample of content from Using R at the Bench: Step-by-Step Analytics for Biologists. Click here for more information or to buy the book. Index A-value, 160 Checking model assumptions, 124 Accuracy, 57 Chi-square test, 85, 97 Adjusted R2, 117, 123 goodness-of-fit, 98 Agglomerative clustering, 150 independence, 99 Alternative Classification, 145–147 hypothesis, 79 error, 147 one-sided, 79 rule, 147 two-sided, 79 Clustering, 145, 148 Analysis of variance, 131 k-means, 153 ANOVA, 113 agglomerative, 150 ANOVA, 113, 131 divisive, 150 assumptions, 140 hierarchical, 150 F-test, 133 partitional, 150, 153 model, 60 Common response, 48, 49 one-way, 132 Complete linkage, 152 two-way, 136 Confidence, 72 Association, 47 Confidence interval, 65, 113 Assumption, 7, 67 computing, 72 ANOVA, 140 interpretation, 67 constant variance, 125 large sample mean, 72 normality, 125 population proportion, 76 Average, 20 small sample mean, 73 Average linkage, 152 Confidence level, 67, 70, 72 Confounding, 48 Bar plot, 24 Constant variance assumption, 125 Bayesian statistics, 145, 176 Contingency table, 19, 60, 98 Bell curve, 33 Continuous variable, 18 Bias, 56 Correlation, 46 Bin, 26 Correlation coefficient, 115, 116 Binomial Critical value, 70 coefficient, 32 computing, 71 distribution, 31 Cumulative probability, 32 Biological replication, 59 Data variation, 58 categorical, 19, 24, 25 BLAST, 110 quantitative, 19, 27, 28 Bootstrap, 64 transformation, 37 Box plot, 28 Dependent variable, 51 Descriptive statistics, 17 Categorical, 60 Design of experiments, 51 data, 19 Deterministic model, 51 variable, 113 Differential expression, 132 Causation, 48 Discrete variable, 17 Cause and effect, 48 Discriminant function, 147 Central Limit Theorem, 39, 66 Discrimination, 147 181 © 2015 Cold Spring Harbor Laboratory Press. -

Stat 427/527: Advanced Data Analysis I
Intro Testing Normality Testing Equal Population Variances Stat 427/527: Advanced Data Analysis I Chapter 4: Checking Assumptions September, 2017 1 / 0 Intro Testing Normality Testing Equal Population Variances Stat 427/527: Advanced Data Analysis I Chapter 4: Checking Assumptions September, 2017 2 / 0 Intro Testing Normality Testing Equal Population Variances I Statistical methods make assumptions about the data collection process and the shape of the population distribution. I If you reject the null hypothesis in a test, then a reasonable conclusion is that the null hypothesis is false, provided all the distributional assumptions made by the test are satisfied. |-If the assumptions are not satisfied then that alone might be the cause of rejecting H0. |-Additionally, if you fail to reject H0, that could be caused solely by failure to satisfy assumptions also. I Hence, you should always check assumptions to the best of your abilities. Three basic assumptions I Data are a random sample. I The population frequency curve is normal. I For the pooled variance two-sample test the population variances are also required to be equal. 2 / 0 Intro Testing Normality Testing Equal Population Variances Testing Normality An informal test of normality can be based on a normal scores plot, sometimes called a rankit plot or a normal probability plot or a normal QQ plot (QQ = quantile-quantile). I plot the quantiles of the data against the quantiles of the normal distribution, or expected normal order statistics (in a standard normal distribution) for a sample with the given number of observations. I The normality assumption is plausible if the plot is fairly linear. -

The Sequential Normal Scores Transformation Arxiv:1705.03496V1
The Sequential Normal Scores Transformation Dr. William J. Conover Department of Mathematics and Statistics, Texas Tech University Dr. V´ıctor G. Tercero-G´omez School of Engineering and Sciences, Tecnologico de Monterrey Dr. Alvaro E. Cordero-Franco Facultad de Ciencias F´ısicoMatem´aticas,Universidad Autonoma de Nuevo Leon May 11, 2017 Abstract The sequential analysis of series often requires nonparametric procedures, where the most powerful ones frequently use rank transformations. Re-ranking the data sequence after each new observation can become too intensive compu- tationally. This led to the idea of sequential ranks, where only the most recent observation is ranked. However, difficulties finding, or approximating, the null distribution of the statistics may have contributed to the lack of popularity of these methods. In this paper, we propose transforming the sequential ranks into sequential normal scores which are independent, and asymptotically stan- dard normal random variables. Thus original methods based on the normality assumption may be used. arXiv:1705.03496v1 [math.ST] 9 May 2017 A novel approach permits the inclusion of a priori information in the form of quantiles. It is developed as a strategy to increase the sensitivity of the scoring statistic. The result is a powerful convenient method to analyze non- normal data sequences. Also, four variations of sequential normal scores are presented using examples from the literature. Researchers and practitioners might find this approach useful to develop nonparametric procedures to address new problems extending the use of parametric procedures when distributional assumptions are not met. These methods are especially useful with large data streams where efficient computational methods are required. -

Expected Normal Order Statistics (Exact and Approximate)
161 AlgorithmAS 177 ExpectedNormal Order Statistics(Exact and Approximate) By J. P. ROYSTON MRC ClinicalResearch Centre, Harrow HA] 3UJ, Middx, UK [ReceivedJanuary 1981. Final revisionJuly 1981] Keywords:RANKITS; EXPECTED NORMAL SCORES; EXPECTED NORMAL ORDER STATISTICS LANGUAGE Fortran66 DESCRIPTIONAND PURPOSE The algorithmsNSCOR1 and NSCOR2 calculate the expectedvalues of normal order statisticsin exactor approximateform respectively. NSCOR2 requireslittle storage and is fast, and hence is suitable for implementationon small computersor certain programmable calculators(HP-67, etc). This is not recommendedfor NSCOR1. Expected normal order statisticsare needed in the calculationof analysisof variancetests of normality,such as W (Shapiro and Wilk, 1965) and W' (Shapiro and Francia, 1972). In a sample of size n the expectedvalue of the rthlargest order statistic is givenby (r- 1)! (n-r)! . -X{l_D(x)}r l{)D(x)}n-r (x) dx, (1) where+(x) = 1/V(27r) exp (-2x2) and 4D(x)= fx OO4(z)dz. Values of E(r,n) accurate to fivedecimal places were obtained by Harter (1961) using numericalintegration, for n = 2(1) 100(25)250(50)400. SubroutineNSCOR1 uses the same techniqueas Harter(1961). Rewritethe integrandin (1) as I(r, n, x) = to(x) exp {loge n! - loge (n - r)! - loge (r - 1)! + (r - 1) tl(x) + (n - r) t2(x) + t3(X)}, where to(X) = X, tl(X) = loge { - @(X)}, t2(x) = lge F(x), t3 = -2{1oge(27c) +x2}. Values of to, t1, t2 and t3 are calculated in the range x = -90(h)90, using the auxiliary subroutineINIT, whichneeds to be called once only.E(r, n) is obtainedby summing the values ofI(r, n, x) and multiplyingthe result by h.