The Sixth Computational Approaches for Cancer Workshop
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

NHSGGC Board Meeting Date of Meeting
BOARD OFFICIAL NHS Greater Glasgow & Paper No. 20/40 Clyde Meeting: NHSGGC Board Meeting Date of Meeting: 25th August 2020 Purpose of Paper: For Noting Classification: Board Official Sponsoring Director: Medical Director and Nurse Director Clinical Governance Annual Report (including the Duty of Candour Report) 2019/2020 Recommendation The Board is asked to note the report, which has been approved for publication by the Clinical and Care Governance Committee. Purpose of Paper The Clinical Governance Annual Report provides a description of the key aspects clinical governance arrangements along with examples of improvement activities. The report contains the Duty of Candour Report, whose publication is legal requirement. The report is made publicly available via the Board’s website. Key Issues to be considered The report provides assurance of the maintenance of the clinical governance arrangements along with our Duty of Candour obligations. The report development and publication is being maintained but the process was compromised by the clinical service focus the pandemic response during the normal reporting period over April and May. 1 BOARD OFFICIAL Any Patient Safety /Patient Experience Issues Data on patient safety and experience is included in the report with assurance of learning and improvements Any Financial Implications from this Paper None specified Any Staffing Implications from this Paper None Specified Any Equality Implications from this Paper None Specified Any Health Inequalities Implications from this Paper None Specified Has a Risk Assessment been carried out for this issue? If yes, please detail the outcome. No Highlight the Corporate Plan priorities to which your paper relates Author – Head of Clinical Governance Date – August 2020 2 BOARD OFFICIAL Clinical Governance Annual Report 2019-2020 NHS GREATER GLASGOW & CLYDE Custodian: Head of Clinical Governance Issue date: July 2020 Status: Draft Version: Draft 3.0 1 BOARD OFFICIAL Section Title Page Front Page 1 Table of Contents 2 1. -

Nursing and the Development of Neonatal Intensive Care Units in the United States, 1955-1982
University of Pennsylvania ScholarlyCommons Publicly Accessible Penn Dissertations 2015 "We Were the Eyes and Ears...": Nursing and the Development of Neonatal intensive Care Units in the United States, 1955-1982. Briana Ralston University of Pennsylvania, [email protected] Follow this and additional works at: https://repository.upenn.edu/edissertations Part of the History Commons, and the Nursing Commons Recommended Citation Ralston, Briana, ""We Were the Eyes and Ears...": Nursing and the Development of Neonatal intensive Care Units in the United States, 1955-1982." (2015). Publicly Accessible Penn Dissertations. 1122. https://repository.upenn.edu/edissertations/1122 This paper is posted at ScholarlyCommons. https://repository.upenn.edu/edissertations/1122 For more information, please contact [email protected]. "We Were the Eyes and Ears...": Nursing and the Development of Neonatal intensive Care Units in the United States, 1955-1982. Abstract ABSTRACT "WE WERE THE EYES AND EARS...": NURSING AND THE DEVELOPMENT OF NEONATAL INTENSIVE CARE UNITS IN THE UNITED STATES, 1955-1982. Briana Ralston, MS, RN Julie Fairman, PhD, RN, FAAN In the 1960s and 1970s, neonatal intensive care units (NICUs) became the standard of care for critically ill newborns in hospitals across the United States. Though work has been done to examine how nurses participated in the development of ICU's for adult populations, scholarship related to the formation of NICUs is sparse. Using historical methodology to examine hospital archival data, oral history interviews, and scholarly literature, this work examines the roles nurses played in the development of NICUs as technological systems between 1955 and 1982 in the United States. -

Notes and Data Sources
Notes and Data Sources Chapter 1 Page 10 Exercise 1.1: From “About six-in-ten Americans support marijuana legal Page 3 Data on climate change due to human activ ization”; available online at http://www ity is from “Public and scientists’ views on .pewresearch.org/fact-tank/2018/01/05 science and society,” Pew Research Cen /americans-support-marijuana-legalization/. ter report by Cary Funk and Lee Rainie, Page 11 The estimates of the census overcount and January 29, 2015; available online at https:// undercount are available online at http:// www.pewresearch.org/science/2015/01/29 www.census.gov/newsroom/releases /chapter-3-attitudes-and-beliefs-on /archives/2010_census/cb12-95.html. -science-and-technology-topics/. This survey contains data on the Page 15 Exercise 1.3: This problem is based on opinions about several other issues of information in the article, “The racial con U.S. adults and AAAS scientists. fidence gap in police performance”; avail able online at http://www.pewsocialtrends Page 3 The MLive.com poll is described in the .org/2016/09/29/the-racial-confidence-gap article “Take our online poll: Should Mich -in-police-performance/. igan legalize marijuana”; available online at http://www.mlive.com/news/kalamazoo Page 15 Exercise 1.5: Richard Westfall, Murray /index.ssf/2014/02/take_our_poll_should Millar, and Mandy Walsh, “Effects of _michigan.html. instructor attractiveness on learning,” Journal of General Psychology, 143 (2016), Page 5 Example 1 is suggested by Maxine pp. 161–171. Pfannkuch and Chris J. Wild, “Statistical thinking and statistical practice: themes Page 17 Exercise 1.12: Jeffrey M. -

Copyright 2017 Laura Atkins
Copyright 2017 Laura Atkins ADAPTABLE LIVES: AGENCY AND ACCOUNTABILITY IN A CANCER CLUSTER TOWN BY LAURA ATKINS DISSERTATION Submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in Sociology in the Graduate College of the University of Illinois at Urbana-Champaign, 2017 Urbana, Illinois Doctoral Committee: Professor Moon-Kie Jung, Chair Professor Assata Zerai Professor Lisa Marie Cacho Professor Norman K. Denzin ABSTRACT This dissertation provides an ethnographic account of the experiences of residents living within the cancer cluster town of Clyde, Ohio, where over 50 children have been diagnosed with or have died of cancers of the brain and central nervous system since the mid-1990s. My entry into the field coincided with the filing of a lawsuit against the town’s largest employer, a Whirlpool Corporation plant, after the discovery of nine feet of toxic PCB sludge at a former community recreational park built by the company. Drawing on in-depth interviews, archival documents, and government reports, I examine systems of power at work within the community that hamper a collective sense of community subpolitics. Using a grounded theoretical approach to analysis informed by risk theory, I discovered that community-level responses to risk echo national logics that promote the concepts of deterrence and avoidance of harm as matters of individual preventive choice. Within a cultural context where efforts towards pinpointing the toxins responsible for the elevated cancer rates in Clyde have failed and there exists an imperative for self-protection that is impossible to achieve, residents experience serious psychosocial and practical conflicts as they adapt to the impact of cancer on their families. -
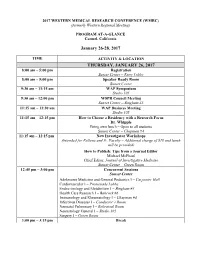
Carmel 2017 Program
2017 WESTERN MEDICAL RESEARCH CONFERENCE (WMRC) (formerly Western Regional Meeting) PROGRAM AT-A-GLANCE Carmel, California January 26-28, 2017 TIME ACTIVITY & LOCATION THURSDAY, JANUARY 26, 2017 8:00 am – 5:00 pm Registration Sunset Center – Entry Lobby 8:00 am – 5:00 pm Speaker Ready Room Sunset Center 9:30 am – 11:15 am WAP Symposium Studio 105 9:30 am – 12:00 pm WSPR Council Meeting Sunset Center – Bingham #3 11:15 am – 11:30 am WAP Business Meeting Studio 105 11:15 am – 12:15 pm How to Choose a Residency with a Research Focus Dr. Whipple Bring own lunch – Open to all students Sunset Center – Chapman #4 11:15 am – 12:15 pm New Investigator Workshops (Intended for Fellows and Jr. Faculty – Additional charge of $10 and lunch will be provided) How to Publish: Tips from a Journal Editor Michael McPhaul Chief Editor, Journal of Investigative Medicine Sunset Center – Green Room 12:45 pm – 3:00 pm Concurrent Sessions Sunset Center Adolescent Medicine and General Pediatrics I – Carpenter Hall Cardiovascular I – Promenade Lobby Endocrinology and Metabolism I – Bingham #3 Health Care Research I – Babcock #6 Immunology and Rheumatology I – Chapman #4 Infectious Diseases I – Conductor’s Room Neonatal Pulmonary I – Rehearsal Room Neonatology General I – Studio 105 Surgery I – Green Room 3:00 pm – 3:15 pm Break 3:15 pm – 5:45 pm Concurrent Sessions Sunset Center Cardiovascular II – Promenade Lobby Community Health I – Carpenter Hall Global Health I – Conductor’s Room Hematology and Oncology I – Green Room Neonatal Pulmonary II – Rehearsal Room Neonatology -

Publications (2005-2020)
Papers from the Sol Goldman Pancreatic Cancer Research Center 2020 1. Abe T, Koi C, Kohi S, Song KB, Tamura K, Macgregor-Das A, Kitaoka N, Chuidian M, Ford M, Dbouk M, Borges M, He J, Burkhart R, Wolfgang CL, Klein AP, Eshleman JR, Hruban RH, Canto MI, Goggins M. Gene Variants That Affect Levels of Circulating Tumor Markers Increase Identification of Patients with Pancreatic Cancer. Clin Gastroenterol Hepatol. 18(5):1161-1169, 2020. 2. Amini N, Rezaee N, Habib JR, Blair A, Beckman RM, Manos L, Cameron JL, Hruban RH, Weiss MJ, Fishman EK, Zaheer A, Lafaro KJ, Burkhart RA, O'Broin Lennon AM, Burns WR, He J, Wolfgang CL. Minimal main pancreatic duct dilatation in small branch duct intraductal papillary mucinous neoplasms associated with high-grade dysplasia or invasive carcinoma. HPB (Oxford). 2020. Online ahead of print. 3. Ardeljan D, Steranka, Liu C,…Hruban R, Boeke J, Fenyo D, Wu P-H, Smogorzewska A, Holland A, Burns K. Cell fitness screens reveal a conflict between LINE-1 retrotransposition and DNA replication. Nat Struct Mol Biol. 27(2):168-178, 2020. 4. Blackford A, Canto M, Klein A, Hruban R, Goggins M. Recent Trends in the Incidence and Survival of Stage 1A Pancreatic Cancer: A Surveillance, Epidemiology, and End Results Analysis. J Natl Cancer Inst. 112(11):1162-1169, 2020. 5. Canto MI, Kerdsirichairat T, Yeo CJ, Hruban RH, Shin EJ, Almario JA, Blackford A, Ford M, Klein AP, Javed AA, Lennon AM, Zaheer A, Kamel IR, Fishman EK, Burkhart R, He J, Makary M, Weiss MJ, Schulick RD, Goggins MG, Wolfgang CL. -

The NIH Catalyst
. Fostering Communication and Collaboration The nihCatalyst A Publication fob NIH Intramural Scientists of of Director Volume 13 Issue 1 a January-February National Institutes Health i Office the e , 2005 Nanomedicine Initiative: Roadmap Recap: A New Pathway to Discovery A Year’s Worth With Evolving Turns and Destinations Of Milestones by Karen Ross by Fran Pollner he aim of nano- raised for her “flawless execu- medicine, one of tion of a very complex process” T nine major NIH by NIH Director Elias Zerhouni, P Roadmap initiatives, is to NIH Roadmap coordinator Dushanka treat disease by intervening Kleinman delivered a first-year at a molecular level. It is a progress report at the De- Roadmap close cousin of nanotech- cember 2004 meeting of the Advi- nology, which is con- sory Committee to the [NIH] Director. cerned with building de- In general, she said, 2004 could vices that are 0.1 (i or less be characterized as a year of seed- in size. (For reference, the ing new tools and technologies for head of a pin is 2,000 jli, a the team science and clinical research typical human cell is ap- of the future. Among the year’s proximately 5 p, and a Intrigued by the Invisible: NEI Director Paul Sieving achievements: large protein complex is (left) and NHGRI tech development guru Jeffery Schloss Establishing four National Centers (right), co-cha irs of the NIH Roadmap Na nomedicine approximately 0.005 p.) for Biomedical Computing (with more Implementation Group flank Richard Fisher, director of Nanomedicines, for ex- , to be funded in 2005) the NEI Corneal Diseases Program and Nanomedicine Launching the Patient-Reported ample, could beof tiny ma- Project team leader Outcomes Measurement Information chines that compensate for System, with seven grants, six to primary the function of defective proteins or very bust systems that operate at the “nano” research centers precisely targeted pharmaceuticals that scale. -

PUBLIC SUBMISSION Posted: January 13, 2011 Tracking No
Page 1 of 1 As of: February 07, 2011 Received: January 03, 2011 Status: Posted PUBLIC SUBMISSION Posted: January 13, 2011 Tracking No. 80bc42e4 Comments Due: February 17, 2011 Submission Type: Web Docket: DOE-HQ-2010-0002 Rulemaking to Amend 10 CFR Part 1021: National Environmental Policy Act Implementing Procedures Comment On: DOE-HQ-2010-0002-0014 National Environmental Policy Act Implementing Procedures Document: DOE-HQ-2010-0002-0019 Comment on FR Doc # 2010-32316 Submitter Information Name: William Kirk Williams Address: 5428 S. Broadwing Way 5428 S. Broadwing Way Boise, Idaho, 83716 Email: [email protected] Fax: (208) 333-9506 General Comment Please do not include "Wind Turbines" and "Solar Potovoltaic" systems among categorical exclusions. Such projects are far too big and consume far too much land to be build without following the EIS process. Wind Turbines kill birds in violation of the Migratory Bird Treaty Act. Without mandatory compliance with EIS no mechanism to assurme BMPs for minimizing bird kills will be in place. Solar Potovoltaic systems have the potential to condemn vast amounts of federal land simultaneously negatively affecting ecosystems of other species, such as sage grouse and economic interests of local communities who might be shut off from existing economic uses of federal lands. One advantage of the EIS processs is considering multiple options for decisions. The use of far too much land is at stake for including both uses as categorical exclusions. file://Z:\2010 CX Rulemaking\01 Comments on Proposed Rule\Comments Received at regulati... 3/9/2011 Page 1 of 1 As of: February 07, 2011 Received: January 03, 2011 Status: Posted PUBLIC SUBMISSION Posted: January 13, 2011 Tracking No. -
UAB 2019 ACADEMIC ANNUAL REPORT Table of Contents
UAB 2019 ACADEMIC ANNUAL REPORT Table of Contents Welcome Letter from Chair .................................................3 High-Impact Research Stories ............................................4 $6.38 million grant to study blood cancers, blood marrow transplantation impact on survivors’ health Researchers awarded $10 million to study acute flaccid myelitis $37.5 million grant will address research of high- priority infections Study aims to discover if hookworm, related intestinal parasites are present in Alabama Black Belt Open source: How data scientist Liz Worthey is bringing precision medicine to the people Department Leadership ......................................................10 Endowed Chairs and Professorships ................................ 11 Pediatric Divisions ................................................................12 Pediatric Residency Program ...................................... 86 Pediatric Fellowship Programs.................................... 89 Kaul Pediatric Research Institute Awards ....................... 93 Dixon Pediatric Fellowship ................................................ 95 Chu Family Educational Scholarship ............................... 96 Russell Cunningham Memorial Research Program ..... 96 Founder’s Fund Grants ........................................................97 Pediatric Research Office .................................................. 98 Office of Faculty Development ...................................... 100 PEDIATRIC SURGICAL SUBSPECIALTIES Pediatric General Surgery -

TITLE Environmental Toxins and Children: Exploring the Risks, Part I
DOCUMENT RESUME ED 336 177 PS 019 535 TITLE Environmental Toxins and Children: Exploring the Risks, Part I. Hearing held in Oakland, California, before the Select Committee on Children, Youth, and Families. House of Representatives, One Hundred First Congress, Second Session. INSTITUTION Congress of the U.S., Washington, DC. House Select Committee on Children, Youth, and Families. PUB DATE 6 Sep 90 NOTE 1S0p.; For part II, hearing held in Washington, D.C., see PS 019 536. Portions contain small print. AVAILABLE FROMSuperintendent of Documents, Congressional Sales Office, U.S. Government Printing Office, Washington, DC 20402 (Stock No. 052-070-06726-8, $5.00). PUB TYPE Legal/Legislative/Regulatory Materials (090) EDRS PRICE MFOI/PCOB Plus Postage. DESCRIPTORS Cancer; *Child Health; *Children; Congenital Impairments; Developmental Disabilities; *Environmental Inflences; Environmental Standards; Lead Poisoning; Migrant Workers; Pesticides; *Poisons; *Pollution; *Public Health IDENTIFIERS California; Child Safety; Passive Smoking; *Risk Assessment ABSTRACT This report contains the proceedings of the first of two hearings that explored the risks of environmental toxins to children. Testimony concerned the special vulnerabil:Lty of chA1dren to toxins; the dangers of lead poisoning; instances ofchildhor>d cancer, birth defects, and developmental problems;passive smoking; child health in the California farm workers' communities; and the relationship between environmental toxins and child healtn as discussed by physicians participating in a Kids and the Environment seminar. A fact sheet describes child vulnerability to environmental toxins; lead poisoning; passive smoking; cancer risk; homepesticide use; and child cancer clusters inCalifornia. A copy of "What's Gotten into Our Children," a report prepared by ChildrenNOW, is included. -

MUC13 Modulated Nanomechanical and Biophysical Responses in Pancreatic Cancer Cells
University of Tennessee Health Science Center UTHSC Digital Commons Theses and Dissertations (ETD) College of Graduate Health Sciences 4-2020 MUC13 Modulated Nanomechanical and Biophysical Responses in Pancreatic Cancer Cells Andrew E. Massey University of Tennessee Health Science Center Follow this and additional works at: https://dc.uthsc.edu/dissertations Part of the Pharmacy and Pharmaceutical Sciences Commons Recommended Citation Massey, Andrew E. (https://orcid.org/0000-0001-7688-5501), "MUC13 Modulated Nanomechanical and Biophysical Responses in Pancreatic Cancer Cells" (2020). Theses and Dissertations (ETD). Paper 511. http://dx.doi.org/10.21007/etd.cghs.2020.0496. This Dissertation is brought to you for free and open access by the College of Graduate Health Sciences at UTHSC Digital Commons. It has been accepted for inclusion in Theses and Dissertations (ETD) by an authorized administrator of UTHSC Digital Commons. For more information, please contact [email protected]. MUC13 Modulated Nanomechanical and Biophysical Responses in Pancreatic Cancer Cells Abstract Pancreatic adenocarcinoma is one of the deadliest forms of cancer. Even with recent advances in diagnostic tools, chemotherapeutic regimens, and biomarkers for earlier detection, it still has dismal survival rates. Part of the reason for this is the inherent difficulty in detecting and eatingtr this disease. Recent findings suggest that the altered expression of mucins, including MUC13, may be useful molecular signatures for early disease diagnosis, chemotherapy response and predicting patient survival. MUC13, a recently identified transmembrane glycoprotein, is normally associated with forming a protective barrier on epithelial tissues. However, its overexpression/aberrant subcellular localization has been associated with cancer, disease aggressiveness, poorer patient prognosis and drug resistance via alterations of multiple oncogenic signaling pathways. -

Best Practices in State and Regional Innovation Initiatives: Competing in the 21St Century
Best Practices in State and Regional Innovation Initiatives: Competing in the 21st Century Charles W. Wessner, Editor Committee on Competing in the 21st Century: Best Practice in State and Regional Innovation Initiatives Board on Science, Technology, and Economic Policy Policy and Global Affairs Copyright © National Academy of Sciences. All rights reserved. Best Practices in State and Regional Innovation Initiatives: Competing in the 21st Century THE NATIONAL ACADEMIES PRESS 500 Fifth Street NW Washington DC 20001 NOTICE: The project that is the subject of this report was approved by the Governing Board of the National Research Council, whose members are drawn from the councils of the National Academy of Sciences, the National Academy of Engineering, and the Institute of Medicine. The members of the committee responsible for the report were chosen for their special competences and with regard for appropriate balance. This study was supported by: Contract/Grant No. DE-DT0000236, TO #28 (base award DE-AM01-04PI45013), between the National Academy of Sciences and the Department of Energy; and Contract/Grant No. N01-OD-4-2139, TO #250 between the National Academy of Sciences and the National Institutes of Health. This report was prepared by the National Academy of Sciences under award number SB134106Z0011, TO# 4 (68059), from the U.S. Department of Commerce, National Institute of Standards and Technology (NIST). This report was prepared by the National Academy of Sciences under award number 99-06- 07543-02 from the Economic Development Administration, U.S. Department of Commerce. The statements, findings, conclusions, and recommendations are those of the author(s) and do not necessarily reflect the views of the National Institute of Standards and Technology, the Economic Development Administration, or the U.S.